Figure 2.
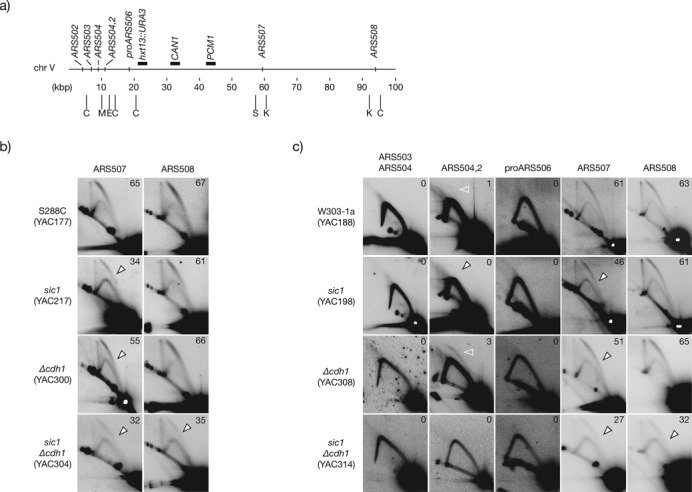
Overall origin firing efficiency at the left arm of chromosome V in CDK-deregulated cells. (a) Map of the first 100 kbp of chromosome V including the relative position of all confirmed and likely origins (according to the OriDB and the SGD). Horizontal black bars above the chromosome denote the relative position of markers for the GCR assay (URA3 and CAN1), and the first essential gene from the left telomere (PCM1) relevant for the GCR assay. Below the chromosome, vertical bars indicate the position of the restriction enzymes employed to digest DNA for 2D gel analysis: C, ClaI; M, MluI; E, EcoRI; S, SacI; K, KpnI. (b) Firing efficiency analysis of ARS507 and ARS508 in S288C control, sic1, Δcdh1 and sic1 Δcdh1, double mutant cells. (c) Firing efficiency of the indicated active, dormant and likely origins at the left arm of chromosome V in W303–1a control and CDK-deregulated cells. Open arrowheads indicate loss of efficiency relative to control cells. Open white arrowheads mark the bubble arc at ARS504.2. See Material and Methods for details of chromosome coordinates and probes employed for each origin.
