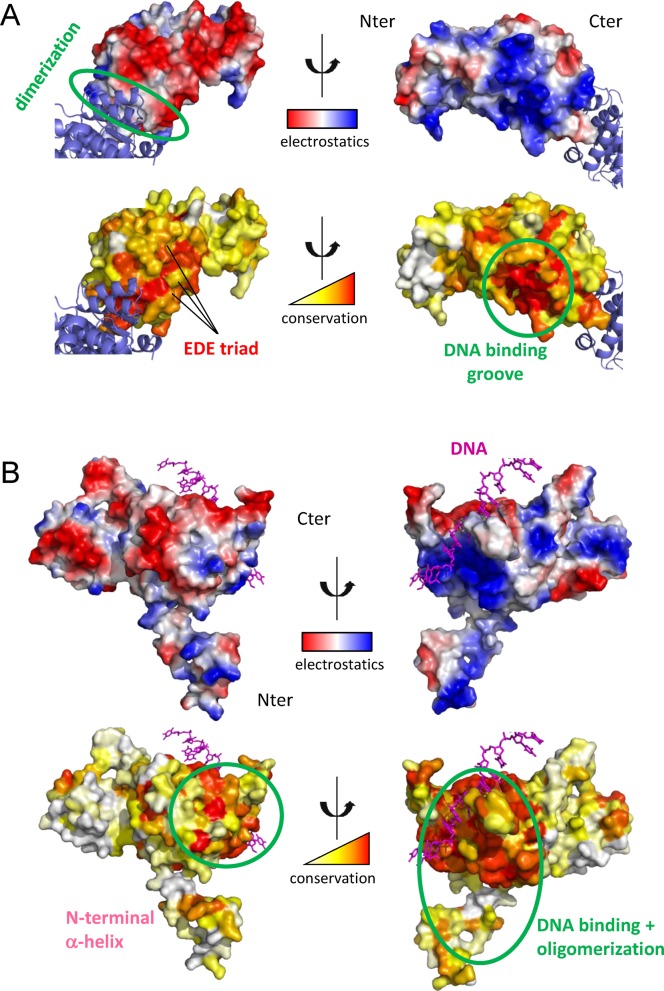
Figure 1.
Structural and evolutionary analysis of the binding partners. Electrostatic surfaces were generated using PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.) (A) SpDprA surface electrostatics (top) and relative evolutionary conservation (bottom). Positively charged regions are in blue and negatively ones in red. The evolutionary conservation scale is colored from white (poorly conserved) to red (highly conserved). The second DprA monomer represented in blue ribbon highlights the dimerization interface that is circled in green. The EDE triad identified in (18) is marked on one monomer. The putative DNA binding groove, characterized by high conservation and highly positive surface, is circled in green. (B) EcRecA surface electrostatics (top) and relative evolutionary conservation (bottom). Single-strand DNA is represented in magenta. The DNA binding groove and the multimerization regions are circled in green.