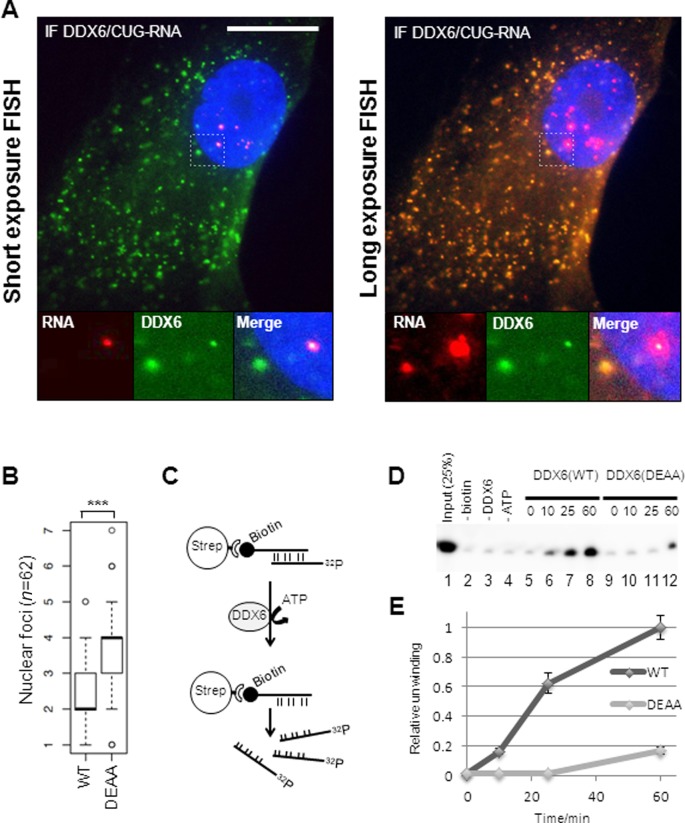
Figure 7.
CUG-foci release to the cytoplasm correlates with co-localization of overexpressed DDX6 and CUG-expanded DMPK-mRNA in DM1 cells. (A) Representative FISH/IF analysis of DDX6 and CUG-expanded DMPK-mRNA in transduced (FLAG-DDX6) DM1 cells with ‘normal’ exposure of the FISH signal (left panel) and ‘longer’ exposure of FISH signal (right panel). Note that nuclear FISH signal is saturated. (B) Boxplot showing quantification of nuclear foci between GFP-DDX6 WT and GFP-DDX6 DEAA expressing cells. Significance was tested using Student's t-test where ‘***’ denotes P < 0.001. (C) RNA helicase assay. A biotinylated 45-mer CUG-oligo was annealed to a radioactively labeled 45-mer CUG-oligo prior to capture on Dynabeads. Immunopurified 3XFLAG-DDX6 from HEK293S cells was incubated with the RNA-duplex and unwinding and release of the radiolabeled strand from dynabeads was monitored. (D) RNA helicase assay. Release of radiolabeled CUG-strand was monitored over time by denaturing acrylamide PAGE. Lane 1: 25% of input RNA. Lane 2: no biotinylated CUG-oligo was added to the annealing reaction prior to capture. Lane 3: no DDX6 was added to the reaction. Lane 4: no ATP was added to reaction. Lanes 5–8: timecourse experiment incubating wild type DDX6 with RNA duplex for 0 min, 10 min, 25 min and 60 min. Lanes 9–12: timecourse experiment incubating DDX6(DEAA) with RNA duplex for 0 min, 10 min, 25 min and 60 min. (E) Quantification of helicase assay using three independent preparations of DDX6 [error bars represent standard deviation (n = 3)].