Figure 6.
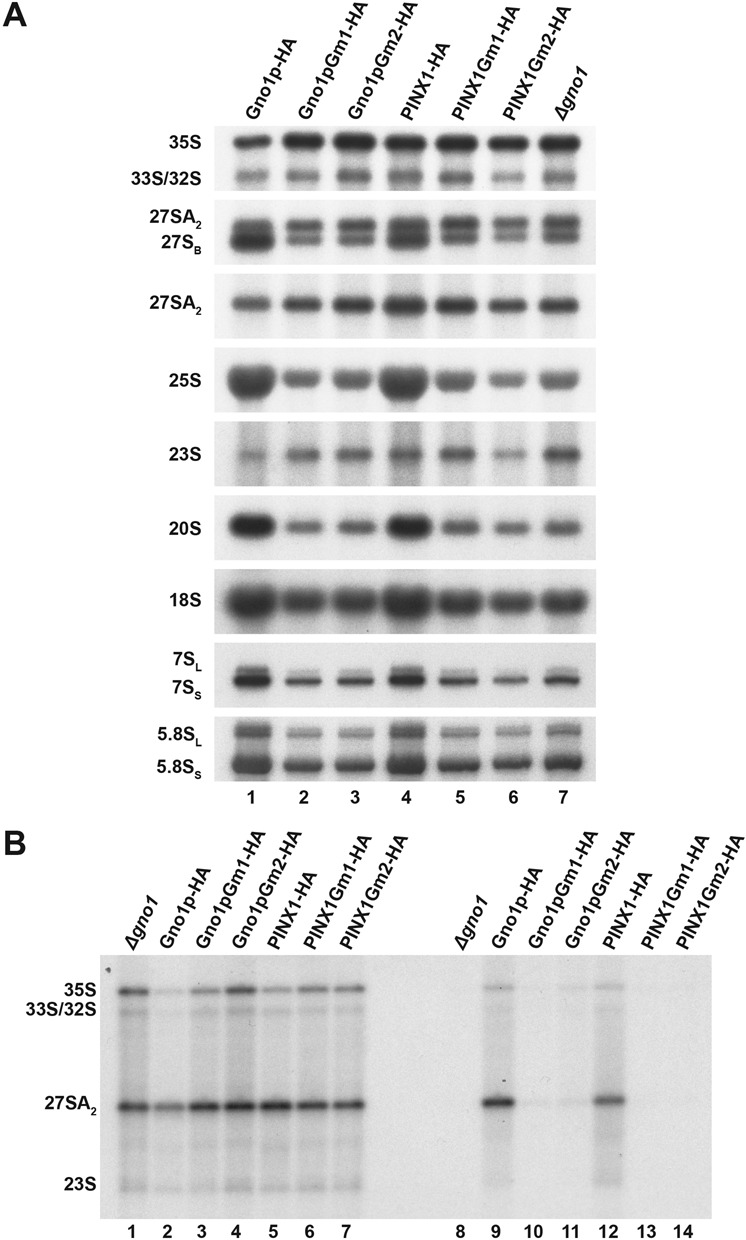
Consequences of alterations to the G-patch domain of Gno1p or PINX1 on ribosome biogenesis in yeast. (A) Northern analysis of rRNAs and pre-rRNAs extracted from Δgno1 cells expressing Gno1p or PINX1 variants featuring amino acid substitutions within their G-patch domain. Total RNAs were extracted from Δgno1 cells transformed with yeast vectors directing expression of wild-type Gno1p-HA (lane 1), Gno1pGm1-HA (lane 2), Gno1pGm2-HA (lane 3), PINX1-HA (lane 4), PINX1Gm1-HA (lane 5), PINX1Gm2-HA (lane 6) or the empty parental vector (lane 7) and analyzed by northern as described in the legend of Figure 4A. (B) Northern analysis of pre-rRNAs co-precipitated with HA-tagged Gno1p, PINX1 or G-patch variants thereof. Immunoprecipitation experiments were performed with an anti-HA matrix using extracts from Δgno1 cells expressing the indicated HA-tagged protein or no tagged protein as control. Input (lanes 1 to 7) and co-precipitated (lanes 8 to 14) pre-rRNAs were analyzed as described in the legend of Figure 5A.
