Figure 2.
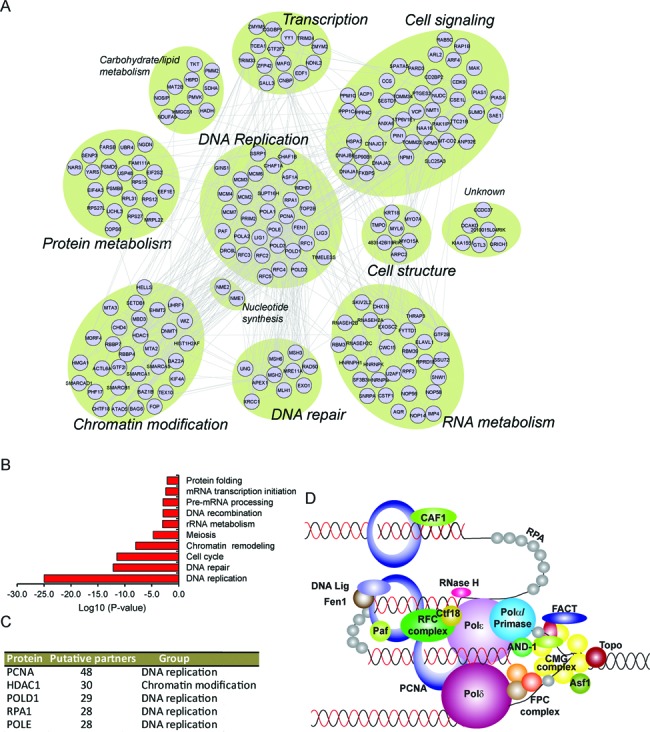
Identification of a protein interaction network associated with nascent DNA in ESCs. (A) The protein–protein interaction network of the proteome as defined by STRING analysis. The topology is organized according to functional classification, as indicated. (B) Gene ontology (GO) analysis of the biological processes of the proteins identified at nascent DNA. The bars indicate the log10 (P-value) for the enriched groups. P-values represent a modified Fisher Exact Test, EASE Score. (C) List of the top five proteins with most interacting partners retrieved from the protein–protein interaction network. (D) Schematic depiction of replication-associated factors that were identified in this study. At least one component of each of the indicated multi-protein complexes was present in our data set. Abbreviations: CMG, for CDC45, MCM2–7 helicases and Gins complex; FPC, for fork-protecting complex (containing TIMELESS, TIPIN and CLASPIN proteins); FACT, for Facilitates chromatin transcription complex (containing SSRP1 and SUPT16H); RFC, for replication factor C complex (containing RFC1–5).
