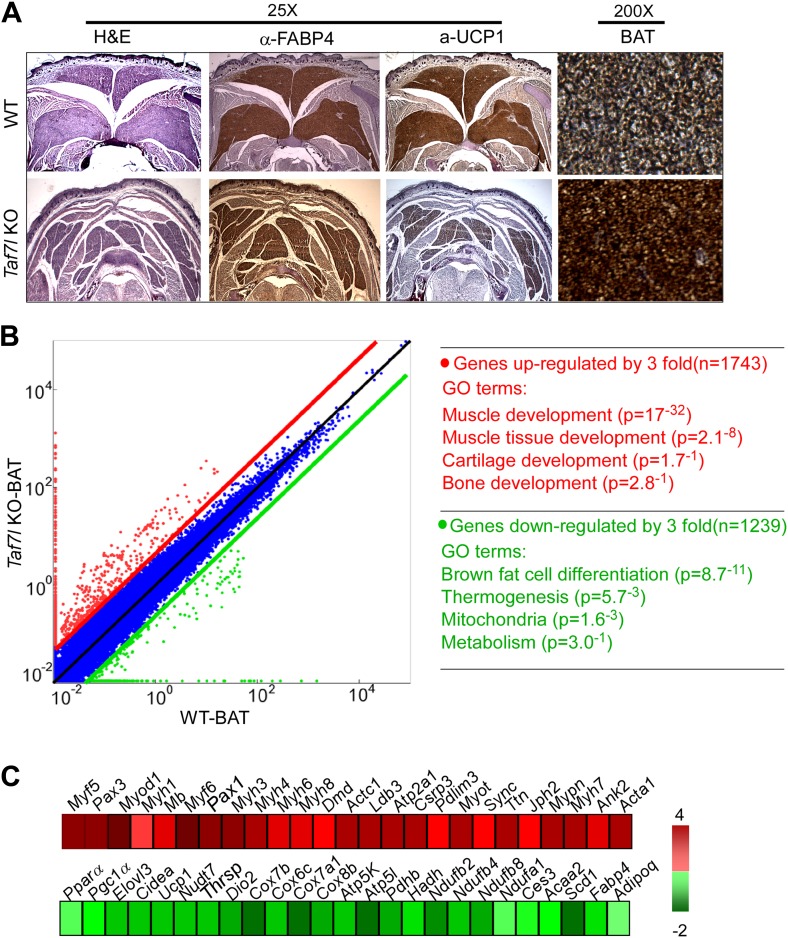
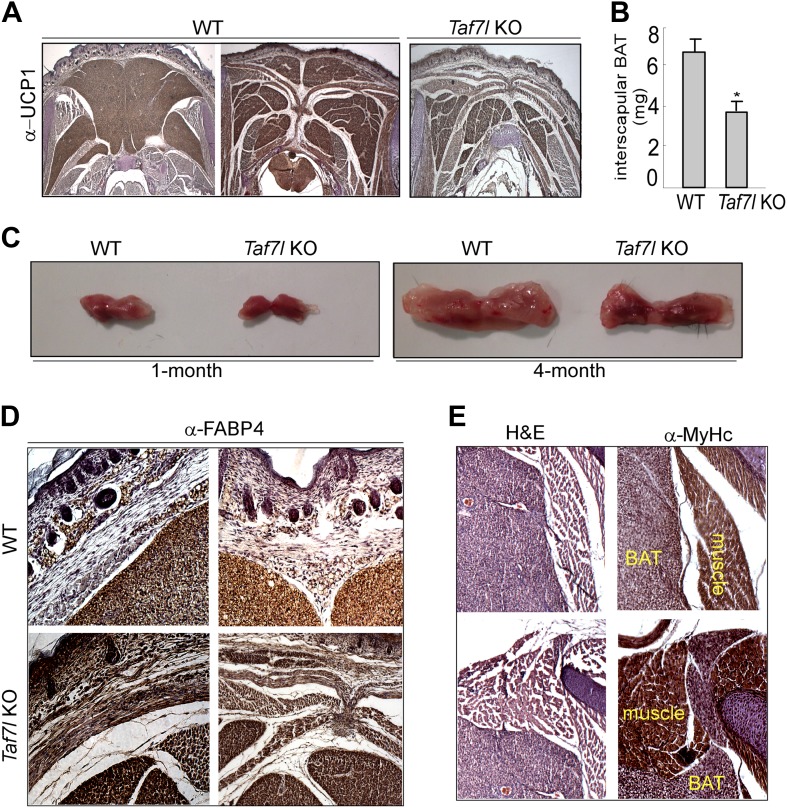
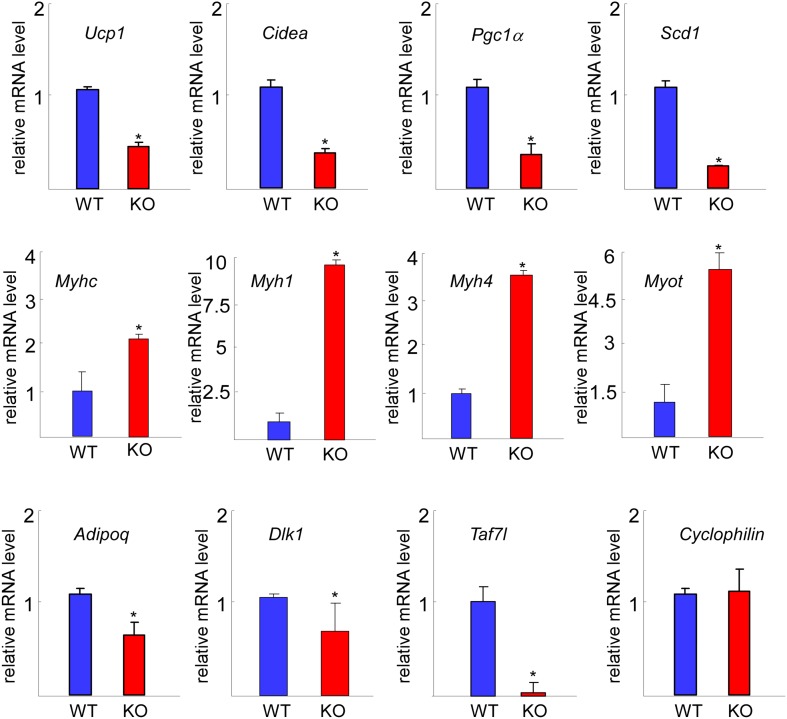
Figure 1. TAF7L is required for proper mouse brown adipose tissue (BAT) formation.
(A) Wild type (WT) and Taf7l knockout (Taf7l KO) interscapular BAT from E18.5 embryos was stained with haematoxylin and eosin (H&E), FABP4, and UCP1 (25X magnification shown). Rightmost panel shows FABP4 staining at 200X magnification. (B) Left panel: scatterplot shows gene expression profile in WT versus Taf7l KO BAT tissue; red dots represent genes up-regulated in Taf7l KO, green dots represent down-regulated genes in Taf7l KO. Right panel: gene ontology analysis shows functional groups of genes changed by at least threefold. (C) Difference in expression of either muscle-specific (top) or brown fat (bottom) genes between WT and Taf7l KO BAT with progressive red and green shades showing degrees of up- and down-regulation, respectively.