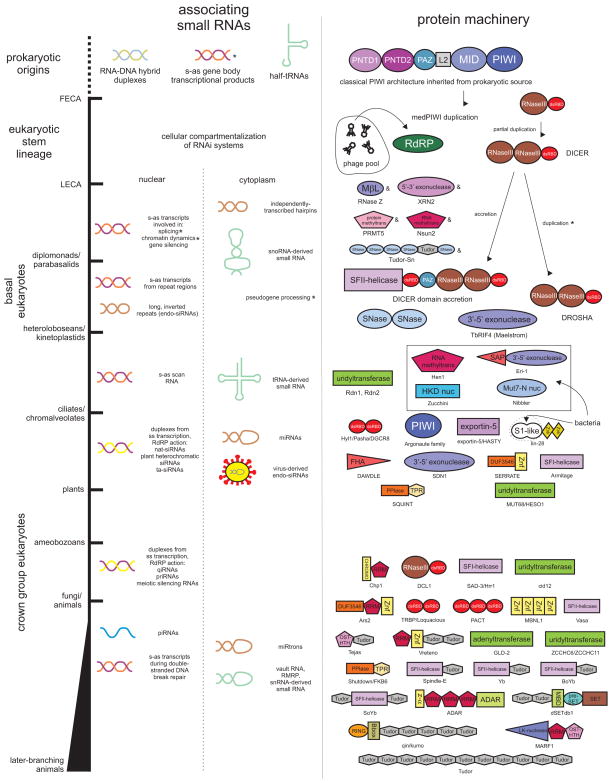
Figure 1.
Temporal diagram depicting emergence of RNA substrates (left column) and core and ancillary protein domains (right column) comprising RNAi systems against the divergence of major eukaryotic lineages labeled to the left. RNAi substrates are divided according to potential for acting in the nuclear and cytoplasmic compartments. Protein domains are depicted by labeled polygonal shapes and are not drawn to scale. Names of the proteins with the depicted architectures are provided below the domains. Inferred origins for several proteins are depicted where appropriate. Affixed asterisks indicate uncertainty in terms of timing of the origins of a RNA substrate or associating protein. Affixed ampersands are indicative of a protein with a deep phylogenetic origin but potential associations with RNAi pathways in early lineages remains unexplored. Abbreviations: PNTD1, PIWI N-terminal domain 1; PNDT2, PIWI N-terminal domain 2, L2, Linker-2 domain; SNase, Staphylcoccal nuclease; nuc, nuclease; Znk, zinc knuckle; FHA, Forkhead-associated; PPlase, peptidyl prolyl-cis/trans-isomerase; Znf, zinc finger; dsRBD, double-stranded RNA-binding domain; HTH, helix-turn-helix; RdRP, RNA-dependent RNA polymerase.