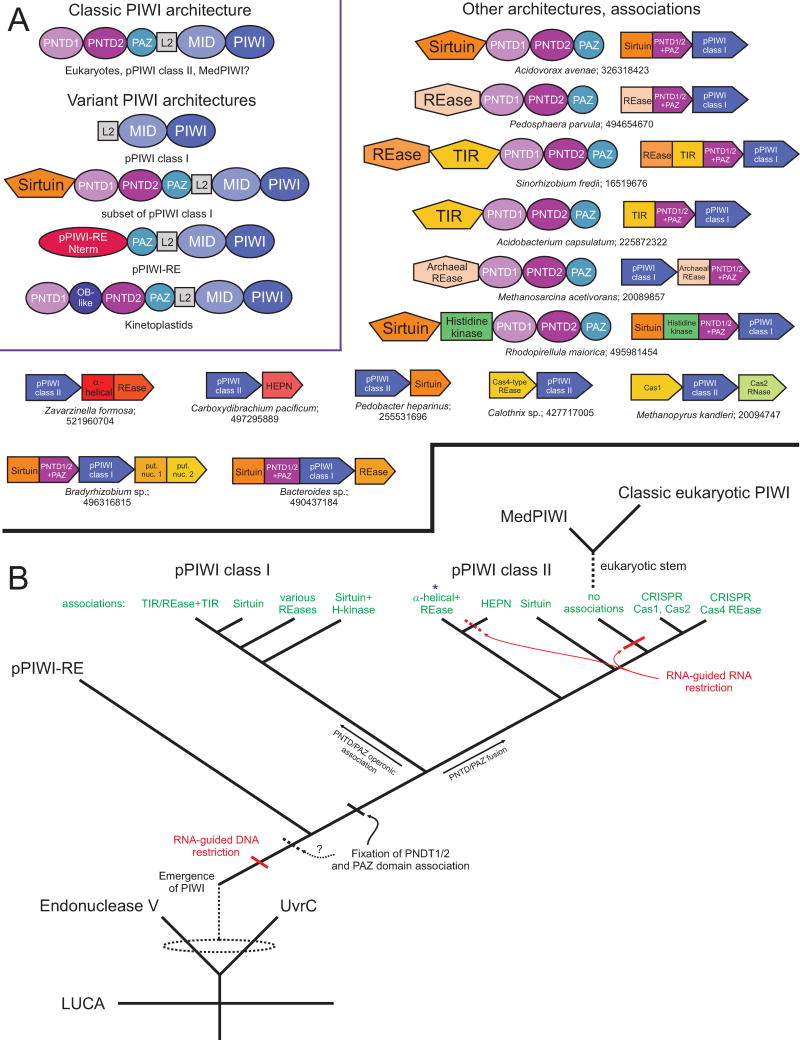
Figure 5.
Genome associations and evolutionary history of the PIWI domain. (A) Domain architectures and conserved gene neighborhoods involving the PIWI domain. The inset, boxed in purple, depicts all known domain architectural themes for the PIWI domain with the phylogenies and/or PIWI families which contain these architectures provided below. Individual domains are depicted as labeled, boxed polygons. Outside of the inset, architectures and conserved gene neighborhoods containing PIWI domains and various nucleases in prokaryotes are provided. Depicted gene neighborhoods are single representatives of a cluster of sequences with the same, conserved neighborhood (for complete lists, see Further Resources). Conserved neighborhoods and domain architectures are labeled by species name and gene identifier (gi) number, separated by a semicolon. (B) Major events in the evolutionary history of the PIWI domain. Emergence of the PIWI domain from an Endonuclease V/UvrC ancestor, the likely cluster of pPIWI domains from which the eukaryotic PIWI versions emerged, and the higher-order relationships of individual PIWI families are depicted. Related groups of pPIWI class I and class II proteins are labeled in green according to the genome associations observed in (A). The group containing the recently-published PIWI-IP seq dataset from R. sphaeroides (α-helical+REase) is denoted with a blue asterisk. Predicted functional shifts for PIWI-centered systems are labeled in red. Dashed lines indicate uncertainty in terms of the origins of a lineage or timing of an event. Abbreviations: REase, Restriction endonuclease; put. nuc., putative nuclease; H-kinase, histidine kinase.