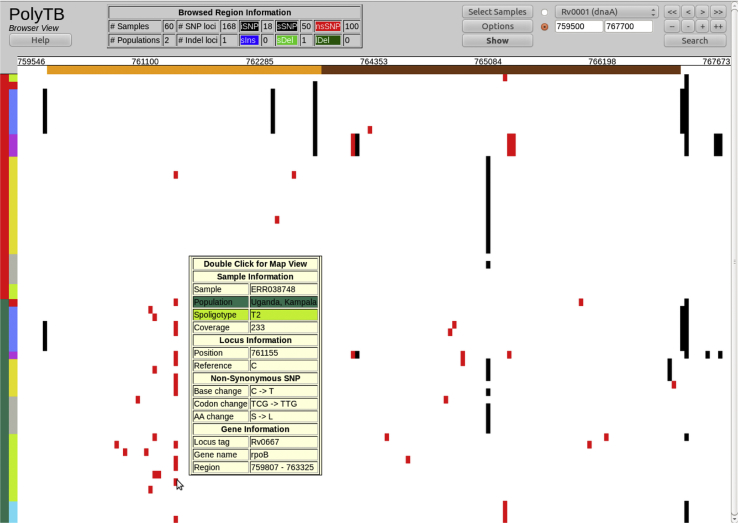
Figure 3.
Polymorphisms at the rpoB-rpoC region associated with rifampicin resistance (Browser View). Genetic variants are shown at the rpoB and rpoC genes, loci known to be associated with rifampicin resistance. Synonymous SNPs (sSNPs) are coloured in black, non-synonymous SNPs (nsSNPs) in red and small insertions and deletions in blue and green, respectively. Cursor movement over variants displays an information box with further annotation including nucleotide, codon and amino acid changes for SNPs; and length and sequence for indels. Locations and Spoligotypes tracks are placed as colour-coded vertical bars at the left hand side of the genomic plot and provide information for samples. Sixty isolates are shown, 30 from Malawi (colour-coded in red in the Location bar) and 30 from Uganda (shown in green). Patterns of SNP difference can be observed when comparing isolates from different populations: Kampala isolates harbour many more nsSNPs at rpoB gene than Malawian isolates. The observed nsSNPs are likely to be the underlying cause of rifampicin resistance (Clark et al., 2013). In fact, rpoB-516 (A → T SNP at 761,110 bp), rpoB-526 (G → T 761,139 bp and A → G 761,140 bp) and rpoB-531 (C → G 761,155 bp) mutations are observed in Ugandan isolates, and correspond to nsSNPs already reported as rifampicin resistance markers [28].