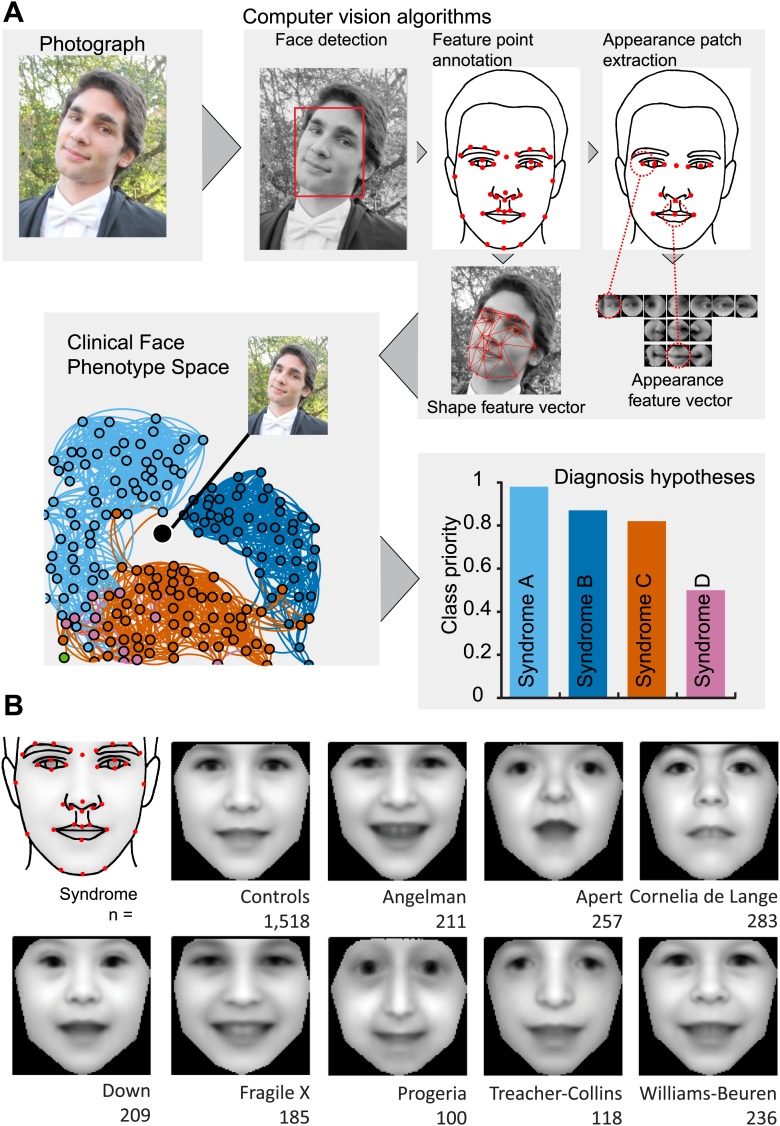
Figure 1. Overview of the computational approach and average faces of syndromes.
(A) A photo is automatically analyzed to detect faces and feature points are placed using computer vision algorithms. Facial feature annotation points delineate the supra-orbital ridge (8 points), the eyes (mid points of the eyelids and eye canthi, 8 points), nose (nasion, tip, ala, subnasale and outer nares, 7 points), mouth (vermilion border lateral and vertical midpoints, 6 points) and the jaw (zygoma mandibular border, gonion, mental protrubance and chin midpoint, 7 points). Shape and Appearance feature vectors are then extracted based on feature points and these determine the photo's location in Clinical Face Phenotype Space (further details on feature points in Figure 1—figure supplement 1). This location is then analyzed in the context of existing points in Clinical Face Phenotype Space to extract phenotype similarities and diagnosis hypotheses (further details on Clinical Face Phenotype Space with simulation examples in Figure 1—figure supplement 2). (B) Average faces of syndromes in the database constructed using AAM models (‘Materials and methods’) and number of individuals which each average face represents. See online version of this manuscript for animated morphing images that show facial features differing between controls and syndromes (Figure 2).