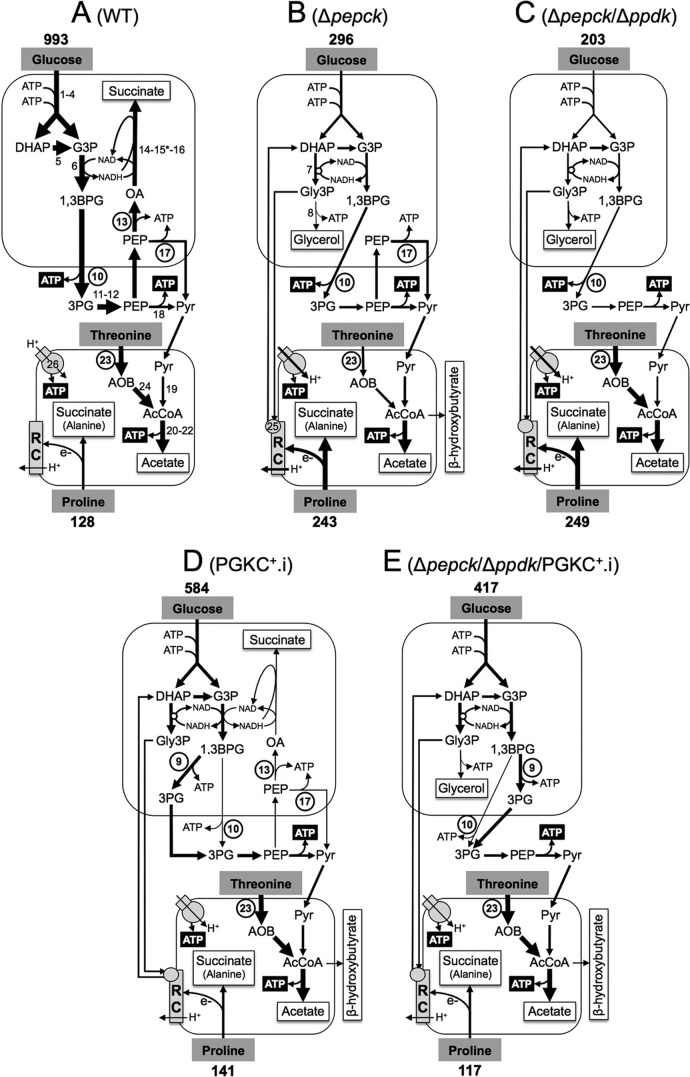
FIGURE 1.
Schematic representation of the intermediary metabolism of procyclic wild-type and mutant cell lines. This figure highlights steps from the glucose metabolism involved in the maintenance of the glycosomal NAD+/NADH and ATP/ADP balances, as well as steps involved in ATP production in the cytosol and mitochondrion from the three main carbon sources used by the procyclic trypanosomes (glucose, proline, and threonine). For simplification, mitochondrial production of succinate from glucose is not shown, as well as the steps of proline degradation and the cytosolic localization of fumarase (step 15*). For enzymatic steps involved in net production of ATP molecules, ATP is represented by white characters on a black background, and the circled step numbers represent enzymes analyzed here. Excreted end products from degradation of glucose, proline and threonine are boxed (succinate and alanine are the main end products excreted from proline metabolism, in the presence or absence of glucose, respectively (14)). The arrow thickness is representative of the measured or estimated metabolic flux through the corresponding branches. The rate of glucose and proline consumption (nmol/h/mg of protein) is indicated above and below the carbon source name, respectively (values from Fig. 3). Abbreviations: AcCoA, acetyl-CoA; AOB, amino oxobutyrate; 1,3BPG, 1,3-biphosphoglycerate; DHAP, dihydroxyacetone phosphate; e−, electrons; G3P, glyceraldehyde 3-phosphate; Gly3P, glycerol 3-phosphate; OA, oxaloacetate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; RC, respiratory chain. Enzymes of glucose of threonine degradation are indicated in panel A, or when mentioned below, in panels B or D: 1, hexokinase; 2, glucose-6-phosphate isomerase; 3, phosphofructokinase; 4, aldolase; 5, triose-phosphate isomerase; 6, glyceraldehyde-3-phosphate dehydrogenase; 7, NADH-dependent glycerol-3-phosphate dehydrogenase (panel B); 8, glycerol kinase (panel B); 9, glycosomal phosphoglycerate kinase (PGKC) (panel D); 10, cytosolic phosphoglycerate kinase (PGKB); 11, phosphoglycerate mutase; 12, enolase; 13, PEPCK; 14, malate dehydrogenase; 15*, cytosolic fumarase (located in the glycosomes for simplification); 16, glycosomal NADH-dependent fumarate reductase; 17, pyruvate phosphate dikinase (PPDK); 18, pyruvate kinase; 19, pyruvate dehydrogenase complex; 20, acetate:succinate CoA-transferase; 21, succinyl-CoA synthetase; 22, acetyl-CoA thioesterase; 23, threonine dehydrogenase (TDH); 24, 2,2-amino-3-ketobutyrate coenzyme A ligase; 25, FAD-dependent glycerol-3-phosphate dehydrogenase (panel B); 26, F0F1-ATP synthase.