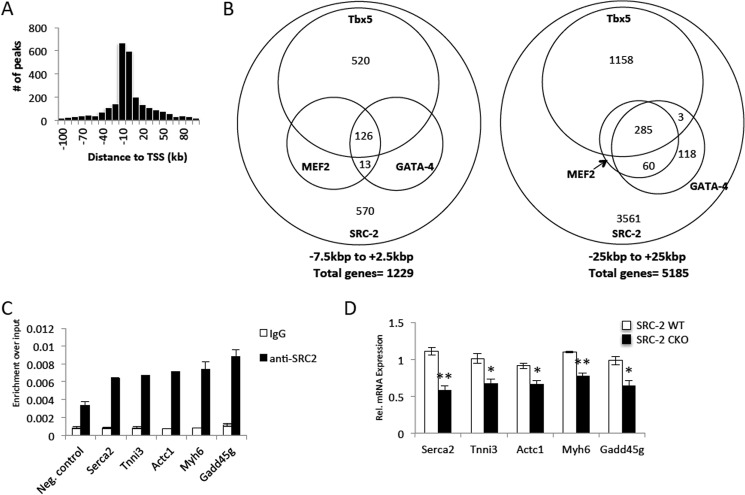
FIGURE 3.
Chromatin occupancy of SRC-2 largely overlaps that of MEF2, GATA-4, and Tbx5. A, analysis of chromatin localization sites for SRC-2 derived from ChIP-Seq analysis of SRC-2 in isolated adult cardiomyocytes as described under “Experimental Procedures” for the distance of each peak relative to the TSS. B, representation of overlap resulting from overlapping binding sites of at least 1 base pair of all binding peaks from the SRC-2 ChIP-Seq overlaid with binding peaks of MEF2, GATA-4, and Tbx5 in HL-1 cells (2). The peaks analyzed were isolated from the indicated regions under each Venn diagram relative to the TSS. C, ChIP analysis as described in Fig. 2C for the indicated target genes. D, qPCR analysis of mRNA expression for the indicated target genes as described in Fig. 1B. Statistical analysis was performed with Student's t test where * = p ≤ 0.05 and ** = p ≤ 0.01.