Figure 2.
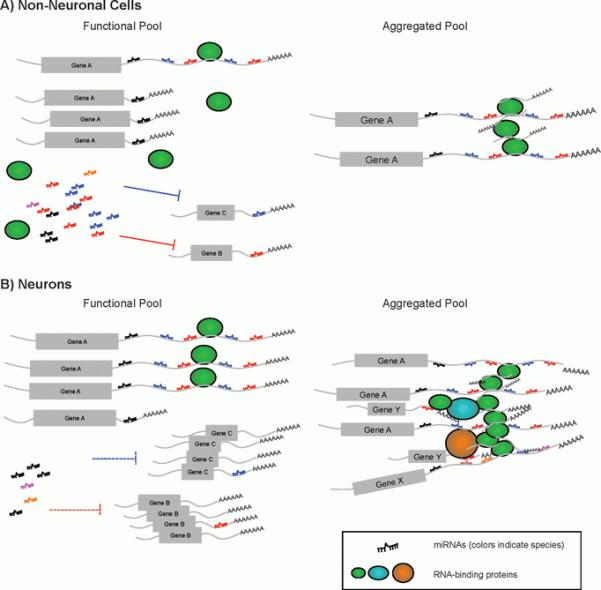
Illustration of potential impact of extended 3′UTRs on miRNA regulation networks and RNP aggregates. A: In non-neuronal cells where dominant 3′UTR isoform of gene A utilizes a proximal polyadenylation signal, the functional pool of miRNAs has an ample quantity of miRNAs (shown as ssRNA in multiple colors), which are sufficient to target gene B and gene C. The transcripts of gene A that are sequestered in the aggregation pool are not significant, and do not interfere with the correspondent miRNA targeting (miRNAs colored black, blue and red). B: In neurons, the long 3′UTR isoform of gene A is drastically upregulated. It may lead to one of two outcomes. First, it serves as a decoy to deplete miRNAs from the functional pool (miRNAs colored black, blue and red), which leads to upregulation of gene B and gene C. Secondly, more gene A transcripts are sequestered to form RNP aggregates. In turn, it depletes other RNA-binding proteins from their functional pool (oval shapes, colored green, cyan, and orange) as well as RNA transcripts of gene X and Y that are associated with these RNA-binding proteins. Collectively, these abnormal aggregates may cause toxicity in pathological conditions.
