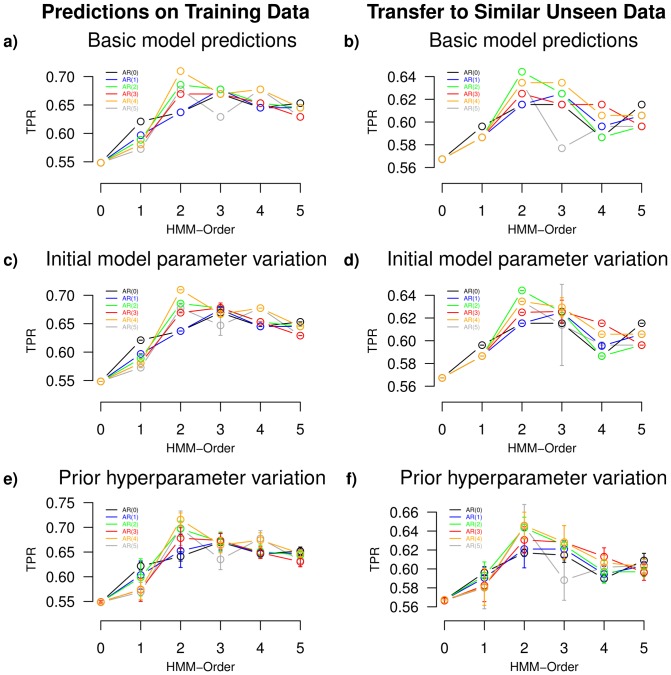
Figure 3. Identification of overexpressed genes with increased copy numbers in breast cancer by different autoregressive HMMs.
Systematic comparison of the identification of overexpressed genes with at least three-fold increased copy numbers by autoregressive HMMs based on breast cancer gene expression profiles from [3]. Each AR( )-HMM(
)-HMM( ) with an emission process of order
) with an emission process of order 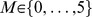 (AR(
(AR(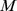 )) in combination with a state-transition process of order
)) in combination with a state-transition process of order 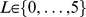 (HMM-Order) is considered. The left column shows the performances reached by autoregressive HMMs trained and applied to fifty percent of the breast cancer gene expression data. The right column shows the performances of these models reached on the remaining unseen fifty percent of the data set. For each model, the identification of candidate genes of overexpression with at least three-fold increased copy numbers is quantified by the true positive rate (TPR) reached at a fixed false positive rate of 5%. Six different scenarios are shown. a) and b) represent performances of the different models with respect to our standard initial model parameter settings. c) and d) represent average performances and corresponding standard deviations reached with respect to systematically changed initial model parameters. e) and f) represent average performances and corresponding standard deviations reached with respect to systematically modified prior hyperparameter settings. The predictions of autoregressive HMMs are generally very robust. Models utilizing a combination of higher-order state-transitions and autoregressive emissions (e.g. AR(
(HMM-Order) is considered. The left column shows the performances reached by autoregressive HMMs trained and applied to fifty percent of the breast cancer gene expression data. The right column shows the performances of these models reached on the remaining unseen fifty percent of the data set. For each model, the identification of candidate genes of overexpression with at least three-fold increased copy numbers is quantified by the true positive rate (TPR) reached at a fixed false positive rate of 5%. Six different scenarios are shown. a) and b) represent performances of the different models with respect to our standard initial model parameter settings. c) and d) represent average performances and corresponding standard deviations reached with respect to systematically changed initial model parameters. e) and f) represent average performances and corresponding standard deviations reached with respect to systematically modified prior hyperparameter settings. The predictions of autoregressive HMMs are generally very robust. Models utilizing a combination of higher-order state-transitions and autoregressive emissions (e.g. AR( )-HMM(
)-HMM( ) and AR(
) and AR( )-HMM(
)-HMM( )) are clearly outperforming the mixture model (AR(
)) are clearly outperforming the mixture model (AR( )-HMM(
)-HMM( )), the standard first-order HMM (AR(
)), the standard first-order HMM (AR( )-HMM(
)-HMM( )), and standard higher-order HMMs (AR(
)), and standard higher-order HMMs (AR( )-HMM(
)-HMM(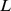 )).
)).