Figure 2.
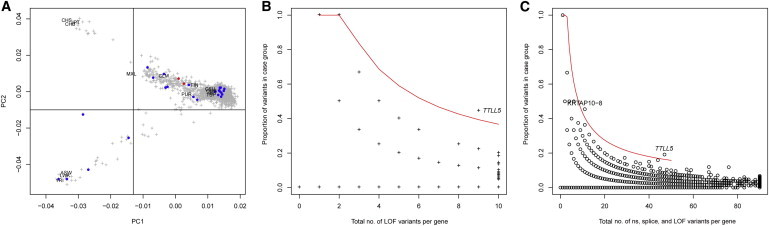
Case-Control Association Results
(A) Principal-component analysis (PCA) of an internal control cohort (“UCL-exomes samples”). PCA was estimated with 1,750 UCL-exomes samples combined with 1,092 samples from diverse ethnic backgrounds; data from the latter were generated as part of the 1000 Genomes Project. Samples selected for the case-control analysis are located in the top right corner of the plot (which includes the samples of European origin). Labels indicate the position of the 1000 Genomes subpopulations. Blue points indicate case samples, and red points indicate the two samples with presumed loss-of-function variants in TTLL5.
(B) Total number of presumed loss-of-function (LOF) alleles in case and control groups (x axis) and the proportion of these alleles in the 23 retinal dystrophy samples (y axis). The area above the red line corresponds to a gene-based p value threshold of p < 10−4.
(C) Same as (B) but for the total number of nonsynonymous (ns) variants (including presumed LOF variants) and splice-site variants (within 5 bp of a splice site). The red line corresponds to the p < 10−5 threshold.
