Abstract
Huntington’s disease (HD) is a fatal dominantly inherited neurodegenerative disorder caused by a CAG repeat expansion leading to an elongated polyglutamine stretch in Huntingtin1. Mutant Huntingtin (mHTT) is ubiquitously expressed but elicits selective cortical and striatal neurodegeneration in HD2. The mechanistic basis for such selective neuronal vulnerability remains unclear. A necessary step towards resolving this enigma is to define the cell types in which mHTT expression is causally linked to the disease pathogenesis. Using a conditional human genomic transgenic mouse model of HD expressing full-length mHTT (BACHD)3, we genetically reduced mHTT expression in striatal, cortical, or both neuronal populations. We show that cortical mHTT reduction in BACHD partially improves motor and psychiatric-like behavioral deficits, but does not improve neurodegeneration, while mHTT reduction in both neuronal populations consistently ameliorates all behavioral deficits and selective brain atrophy in this HD model. Furthermore, mHTT reduction in cortical or striatal neurons partially ameliorates cortico-striatal synaptic deficits, while further restoration of striatal synaptic function is achieved by mHTT reduction in both neuronal cell types. Our study demonstrates distinct, but interacting roles of cortical and striatal mHTT in disease pathogenesis and suggests that optimal HD therapeutics may require targeting mHTT in both cortical and striatal neurons.
Huntington’s disease (HD) is characterized by progressive motor, psychiatric and cognitive deficits with an inexorable and fatal disease course4. HD postmortem brains show degeneration of the majority of striatal medium spiny neurons (MSNs) and, to a lesser extent, cortical pyramidal neurons (CPNs)2. Recent imaging studies suggest early and progressive loss of axons emanating from cortical neurons, including those projecting to the striatum, suggesting early dysfunction of cortico-striatal connectivity in HD5. Currently, there is no therapy to prevent or slow the pathogenesis of HD.
Landmark genetic research has led to the identification of the causal genetic mutation for HD as a CAG repeat expansion translated into an expanded polyglutamine (polyQ) repeat in Huntingtin (HTT) protein1. HTT is ubiquitously expressed and capable of interacting with hundreds of brain proteins6,7. Among the fundamental but unsolved questions in HD pathogenesis are what cell types mutant HTT (mHTT) primarily targets to elicit the disease, whether the vulnerable cortical and striatal neurons are affected through cell-autonomous or non-cell-autonomous mHTT toxicities8, and whether disease processes in these brain regions are causally linked9. Answers to these questions are highly relevant to advancing therapeutics that directly target HTT10–13.
To gain insights into such questions, we developed a conditional Bacterial Artificial Chromosome (BAC) transgenic mouse model of HD (BACHD) that expresses human full-length mHTT (fl-mHTT) with 97Q3. BACHD mice exhibit progressive motor and psychiatric-like behavioral deficits and selective cortical and striatal atrophy but lack overt striatal neuronal loss, hence partially recapitulating the HD phenotypes3,14. BACHD is the first conditional human genomic transgenic mouse model for HD featuring strategically placed LoxP sites flanking mHTT-exon1 with the polyQ repeat, and hence the expression of fl-mHTT can be genetically reduced in Cre-expressing cell lineages (Fig. 1a).
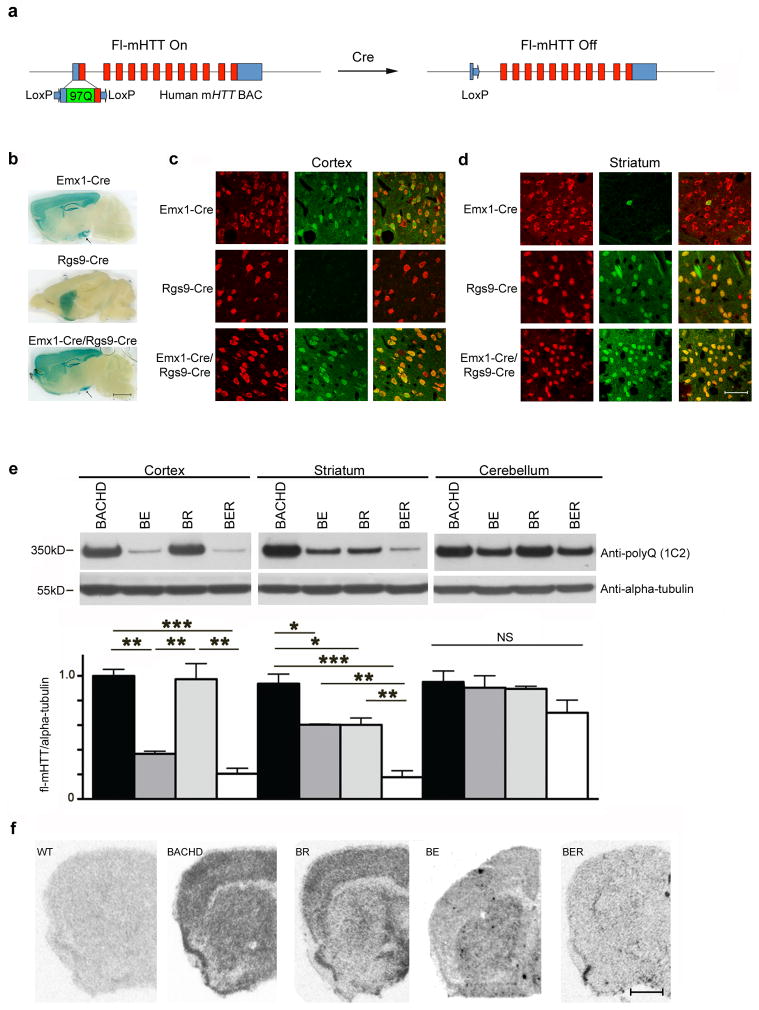
Figure 1. Genetic reduction of full-length human mHTT in cortical, striatal or both neuronal populations in BACHD mice.
(a) Schematic representation of Cre-mediated genetic reduction of mHTT in BACHD mice. Mutant HTT Exon-1 in the BACHD transgene is flanked by two LoxP sites, so fl-mHTT levels can be genetically reduced in cell types expressing Cre. (b) Histochemical staining for β-galactosidase to illustrate the Cre-mediated recombination patterns in double transgenic mice containing the Rosa26-LacZ reporter and one of the Cre mouse lines. Black arrows point to the ventral hippocampus, where Cre recombinase is expressed in Emx1-Cre and Emx1-Cre/Rgs9-Cre lineages. Scale bar = 2 mm. (c,d) Cre recombinase expression patterns in cortex (c) and striatum (d) of Emx1-Cre, Rgs9-Cre and Emx1-Cre/Rgs9-Cre mice crossed with Ai3 Cre reporter mice (mediating Cre-dependent YFP expression)20. Each panel is a composite that shows Cre activity (YFP signals in green) and neuronal staining (NeuN immunofluorescence in red). Scale bar = 100 μm. (e) Western blot with 1C2 (specific to expanded polyglutamine epitope) to detect mHTT but not WT murine Htt in BACHD mice. Subsequent quantitation reveals fl-mHTT protein levels in the cortex, striatum, and cerebellum of BACHD, BE, BR and BER mice (n = 4 per genotype; ***P < 0.001, **P < 0.01, *P < 0.05, one-way ANOVA followed by LSD post hoc test). (f) In situ hybridization with human mHTT exon-1 specific riboprobe reveals selective reduction of mHTT transcripts in the cortex, striatum or both regions in BE, BR and BER brains, respectively. Scale bar = 1 mm.
To address whether mHTT expression in MSNs, CPNs, or both could be responsible for aspects of disease phenotypes in BACHD, we first chose Cre mouse lines (Rgs9-Cre and Emx1-Cre) that have specificity to these neuronal populations15–18. Using distinct reporter mice with Cre-dependent expression of LacZ19 or YFP (Ai3 mice)20, we confirmed that Rgs9-Cre selectively activates Cre-dependent gene expression in striatal MSNs (Fig. 1b–d) but not in other brain regions (Supplementary Fig. 1a). Consistent with prior studies, Emx1-Cre is active in the CPNs in the cortex and hippocampus, a subset of neurons in the amygdala, and only a few neurons in the striatum (about 4%) and cerebellum (Fig. 1c,d and Supplementary Fig. 1a). In the thalamus, Emx1-Cre/Ai3 mice showed YFP signal in the neuropil (likely in the axon terminals of the CPNs), but not in the neuronal cell bodies (Supplementary Fig. 1a,b). Thus, our study confirmed that Emx1-Cre is a relatively cortex-specific Cre mouse line15,17,18,21. Finally, we showed that Emx1-Cre/Rgs9-Cre double transgenic mice efficiently drive Cre-dependent gene expression in both CPNs and MSNs (Fig. 1b–d), but very few other neuronal populations in the brain (Supplementary Fig. 1a).
To generate a BACHD model series with genetic reduction of mHTT in MSNs, CPNs, or both, we crossed BACHD with Emx1-Cre/Rgs9-Cre double transgenic mice in the FvB/NJ inbred background. A cohort of BACHD, BACHD/Emx1-Cre (BE), BACHD/Rgs9-Cre (BR), BACHD/Emx1-Cre/Rgs9-Cre (BER) mice were generated and used to confirm selective reduction of mHTT mRNA and protein in distinct brain regions (Fig. 1e,f). Consistent with MSNs constituting a subset of the cells in the striatum (about 68%)22, we found that BR mice have selective partial reduction of mHTT protein (about 50%) in the striatum, but not in the cortex or cerebellum (Fig. 1e). BER mice have significant (about 80% reduction) of mHTT compared to BACHD in both cortex and striatum, but not in the cerebellum (Fig. 1e). Interestingly, BE mice have about 70% reduction of mHTT protein in the cortex, but also about 40% reduction of mHTT protein in the striatum (Fig. 1e). Since in situ hybridization reveals that mHTT transcripts show selective reduction in BE cortex (Fig. 1f), and BER Western blots showed further significant mHTT protein reduction in the striatum compared to either BE or BR littermates (i.e. mHTT protein is reduced in different “compartments” in BE and BR; Fig. 1e), we conclude that the decrease of the striatal mHTT protein level in BE is due to reduction of mHTT protein that is synthesized in the cortical neurons and transported into the striatum within the corticostriatal and other corticofugal axons. Finally, Western blot analyses also demonstrate that Cre-mediated mHTT reduction in BACHD mice does not create any aberrant mHTT N-terminal fragments (Supplementary Fig. 2), and hence the phenotypic differences between BE, BR and BER can be interpreted in the context of fl-mHTT genetic reduction in the specific cortical or striatal neuronal cell types.
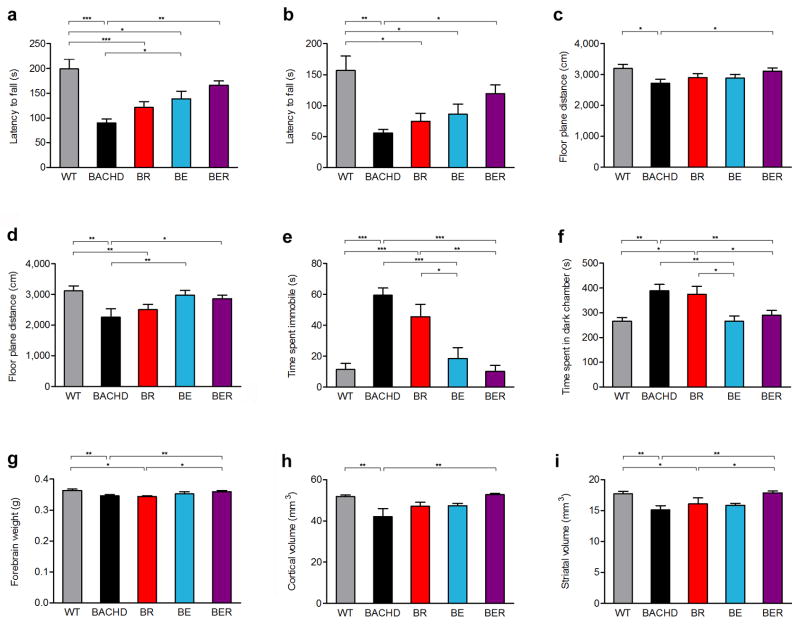
We next tested whether age-dependent motor and psychiatric-like behavioral deficits in BACHD can be improved via genetic reduction of mHTT in cortical or striatal neurons. We characterized a cohort of BACHD, BE, BR, BER and wildtype (WT) mice at 2-months (2m), 6-months (6m) and 12-months (12m) using Rotarod (motor performance) and Open-field (locomotion) tests, which are reliable assays of motor impairment in BACHD3,14. BACHD mice show the largest progression of Rotarod deficit between 2m and 6m of age, while the progression is smaller albeit statistically significant between 6m and 12m (Fig. 2a–d, Supplementary Figure 3) 3,14. Compared to WT mice, BR mice show significant Rotarod impairment at 6m and 12m, and reduced locomotion at 12m (Fig. 2a–d, Supplementary Table 1). Interestingly, although BE mice are still motorically impaired compared to WT, they do show significant, albeit partial amelioration of Rotarod performance at 6m and Open-field at 12m compared to BACHD (Fig. 2a, d, Supplementary Table 1). Importantly, BER mice show consistent and significant improvement of both motor phenotypes compared to BACHD at both testing ages (Fig. 2a–d, Supplementary Table 1).
Figure 2. Differential amelioration of behavioral deficits and selective neurodegenerative pathology in BACHD mice with genetic mHTT reduction in cortical or striatal neurons.
A cohort of BACHD, BE, BR, BER, and WT littermate mice in FvB/NJ background were tested for longitudinal behavioral deficits. Two motor tests were used: Accelerating Rotarod at 6m (a) and 12m (b) of ages, and spontaneous locomotion in an Automated Open-Field Test at 6m (c) and 12m (d) of ages. Moreover, the mice were also tested for psychiatric-like behavioral deficits using two tests: depression-like behaviors in the Forced Swimming Test (e) and anxiety-like behaviors in the Light-Dark Box Exploration Test (f). Mice of different genotypes at 12m of age were used to quantify forebrain weights (g) in addition to unbiased stereological measurement of cortical (h) and striatal (i) volumes. One-way ANOVA followed by LSD post hoc test was applied for all behavioral studies. *P < 0.05, **P < 0.01, ***P < 0.001, values represent means ± SEM. For 6-month Rotarod and Open Field Tests, n = 14, 14, 15, 13 and 15 for WT, BACHD, BR, BE and BER, respectively; for all other behavioral tests, n = 12 per genotype; n = 8 per genotype in all pathological studies (more detailed statistics in Supplementary Table 1).
BACHD mice exhibit psychiatric-like behavioral deficits including anxiety-like behavior in light-dark box exploration and depression-like behavior in forced swimming test (Fig. 2e,f)14. Interestingly, mHTT reduction in both BE and BER mice significantly improves both psychiatric-like behavioral deficits compared to BACHD, while BR mice are still impaired compared to WT mice. The psychiatric-like behavioral deficits in BACHD and BR mice cannot be simply attributed to motor impairment that interferes with testing, since the initial latency to enter the dark compartment from the well-lit compartment (a measure of locomotor ability to perform the task) is not significantly different among the genotypes (Supplementary Fig. 4).
We next addressed where mHTT expression is critical for selective neurodegeneration, and whether non-cell-autonomous disease mechanisms could contribute to the disease pathogenesis. Our prior study using a conditional mHTT-exon1 model showed that non-cell-autonomous mHTT fragment toxicities likely contribute to neuronal toxicities, but the precise cellular origins for such toxicities remain unclear15. BACHD as a human genomic transgenic mouse model is particularly suitable to address this question due to its relatively accurate construct validity23 and manifestation of selective brain atrophy that partially mimics HD3. BACHD mice at 12m show selective forebrain weight loss and stereologically-measured cortical and striatal volume loss (Fig. 2g–i). Importantly, the cerebellar weights do not significantly differ among all the genotypes (Supplementary Fig. 5), supporting HD-like neurodegenerative pathology in this model3. In our cohort of HD mice with brain-regional-specific fl-mHTT genetic reduction, only BER mice consistently show significant amelioration of the forebrain weight loss (Fig. 2g) and cortical and striatal volume loss compared to BACHD (Fig. 2h,i). BR mice at 12m still show significant brain atrophy in some (i.e. forebrain weight) but not all the readouts (Fig. 2g–i, Supplementary Table 1). Interestingly, BE mice do not show significant difference in all three brain atrophy phenotypes from neither BACHD or WT, suggesting that cortical mHTT reduction alone can exert behavioral benefit without significant ameliorate neurodegeneation phenotypes in BACHD (Fig. 2g–i, Supplementary Table 1).
Our study reveals a remarkably consistent finding regarding the neuronal targets in which mHTT synthesis significantly contributes to disease pathogenesis (Table 1): First, genetic reduction of fl-mHTT in both cortical and striatal neurons in BER mice consistently improves all disease phenotypes measured in BACHD. Second, genetic reduction of mHTT in the cortical neurons in BE mice showed significant amelioration of psychiatric-like behavioral deficits, and partial improvement in motor deficits, suggesting that mHTT synthesized in the cortical neurons play an important role in the pathogenesis of behavioral deficits in HD mice. Finally, although we did not show any significant improvement of the behavioral deficits and neuropathology in BR mice compared to BACHD (Table 1), these data should be interpreted with caution, since our power analyses (Supplementary Table 1) suggest that a much larger sample size would be needed to conclusively show the absence of phenotypic improvement in BR mice.
Table 1. Summary of the behavioral and neuropathological phenotypic improvement of BE, BR and BER mice compared to BACHD mice.
Percentage of improvement for a given phenotype represents the absolute mean value of the difference between BE/BR/BER and BACHD divided by the absolute mean value of the difference between BACHD and WT.
| BACHD Phenotypes | Significant Disease Phenotype Amelioration (Percentage of Improvement) | ||
|---|---|---|---|
| BR | BE | BER | |
| 1. Motor Deficits | |||
| Rotarod (6M) | NS (29%) | * (45%) | ** (70%) |
| Rotarod (12M) | NS (19%) | NS (30%) | * (63%) |
| Spontaneous Locomotion (6M) | NS (39%) | NS (35%) | * (82%) |
| Spontaneous Locomotion (12M) | NS (30%) | ** (83%) | * (71%) |
| 2. Psychiatric-like Behaviors | |||
| Anxiety (Light-Dark Box) | NS (29%) | *** (85%) | *** (100%) |
| Depression-like (Forced Swim Test) | NS (12%) | ** (100%) | ** (80%) |
| 3. Neurodegeneration | |||
| Forebrain Weight | NS (−17%) | NS (32%) | * (81%) |
| Cortical Volume Loss | NS (51%) | NS (54%) | ** (100%) |
| Striatal Volume Loss | NS (36%) | NS (27%) | ** (100%) |
NS: Not Significant
p<0.05,
p<0.01,
p<0.001
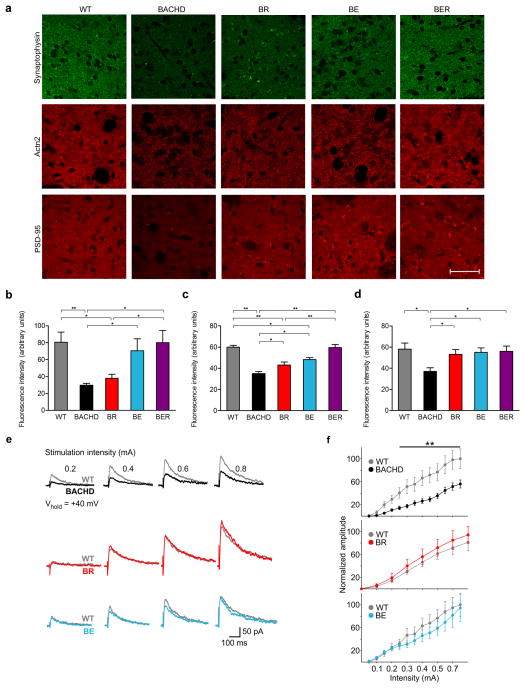
To further explore mechanisms underlying the differential rescue of disease phenotypes in our BACHD model series, we hypothesized that genetic reduction of mHTT in cortical or striatal neurons may have differential impact on cortico-striatal synaptic communication, which is known to be affected in HD24. HD mouse models often recapitulate the loss of synaptic marker proteins, such as the presynaptic protein synaptophysin and postsynaptic proteins Actinin 2 (Actn2) and PSD-9524–28. Interestingly, both Actn2 and PSD-95 are in the postsynaptic density (PSD) of MSNs and functionally interact with N-Methyl-D-Aspartate (NMDA) receptors29–31. The reduction of PSD-95 has been implicated in NMDA receptor dysfunction in HD24,32. By quantifying the immunofluorescence staining signals for these synaptic marker proteins in the dorsolateral striatal sections from 12m old mice, we show that BACHD mice exhibit significant reduction of all three synaptic marker proteins compared to WT mice (Fig. 3a–d). Interestingly, BR mice showed a significant increase of signals for both Actn2 and PSD-95 compared to BACHD, but with Actn2 levels still lower than WT and BER (Fig. 3c). Moreover, BR mice still exhibit significant loss of the presynaptic marker protein synaptophysin compared to WT (Fig. 3b). This result suggests that reduction of PSD marker proteins in MSNs is at least in part due to the cell-autonomous toxicities of mHTT within MSNs. In BE mice, we found a significant improvement of synaptophysin levels in the striatum (Fig. 3b), and surprisingly also significant amelioration of both Actn2 and PSD-95 deficits compared to BACHD, but with the level of Actn2 still lower than WT (Fig. 3c,d). These results suggest that mHTT reduction in CPNs can not only lead to cell-autonomous improvement of presynaptic marker protein levels in striatum but originated from the CPNs, but also exert non-cell-autonomous benefit on the postsynaptic marker protein expression in the MSNs. Finally, BER mice show the most consistent improvement of both presynaptic and postsynaptic marker proteins compared to BACHD, and also show significant improvement in Actn2 compared to BR (Fig. 3b–d). The latter finding suggests that, at least for this PSD protein, the reduction of mHTT in both cortical and striatal neurons elicits further disease-reduction benefit than mHTT reduction in the striatal MSNs alone.
Figure 3. Differential improvement of the levels of striatal presynaptic and postsynaptic marker proteins, as well as NMDA receptor-mediated synaptic currents in BACHD mice with genetic reduction of mHTT in cortical or striatal neurons.
(a) The dorsolateral striatal sections from WT, BACHD, BE, BR and BER were immunostained for a presynaptic protein (synaptophysin) or two postsynaptic proteins (Actn2 and PSD-95). Scale bar = 100 μm. (b–d) Quantitation of relative fluorescence intensities reveals the levels for synaptophysin (b; n = 7 per genotype), Actn2 (c; BR, n = 10; all other genotypes, n = 7) and PSD-95 (d, n = 7 per genotype). *P < 0.05, **P < 0.01, values represent means ± SEM. More detailed statistics are presented in Supplementary Table 1. To evaluate cortical-striatal NMDA receptor function, evoked synaptic NMDA receptor-mediated currents in the MSNs of WT, BACHD, BE and BR mice were recorded. (e) The representative traces of NMDA receptor-mediated currents in the MSNs using patch-clamp recording in the presence of BIC and CNQX (n = 9–16 cells per group). All recordings were performed in transgenic mice and WT littermates. The ages of the mice were 13.3m, 14.2m and 15.7m for BACHD, BE and BR mice, respectively. (f) The quantification of normalized amplitude of NMDA receptor-mediated currents in striatal MSNs from BACHD, BR and BE mice compared to those from WT mice. Two way ANOVAs followed by Bonferroni post hoc tests. Values represent means ± SEM, **P < 0.01.
Since both cortical or striatal mHTT reduction can significantly improve the postsynaptic proteins known to regulate NMDA receptor function29–31, we hypothesized that deficits in NMDA-mediated synaptic currents, if found in BACHD, could be ameliorated in BE or BR mice. To test this hypothesis, we examined striatal slices from BACHD, BE, BR and WT mice at a relatively advanced disease stage (13m–15m) for evoked synaptic NMDA currents. We found significantly impaired evoked synaptic NMDA currents in BACHD MSNs compared to those in WT (Fig. 3e,f). Importantly, both BE and BR mice no longer exhibit such deficits compared to respective WT littermates (Fig. 3e,f). To further strengthen the observation that cortical mHTT reduction can ameliorate striatal synaptic toxicities, we examined additional synaptic properties of MSNs in striatal slices from 12m old BE and BACHD mice. We found that BACHD MSNs exhibit a significant reduction of spontaneous excitatory post-synaptic currents (sEPSCs, frequencies at 5–15 pA) and a significant increase of spontaneous inhibitory postsynaptic currents (sIPSCs) compared to WT MSNs. These deficits are significantly ameliorated in MSNs of BE mice (Supplementary Fig. 6). Together, we conclude that mHTT in both striatal and cortical neurons contributes to the molecular and physiological deficits found in the synapses of striatal MSNs. Moreover, we demonstrated that mHTT synthesized in cortical neurons exerts non-cell-autonomous toxicities that contribute to synaptic pathogenesis within striatal MSNs.
In summary, our study demonstrates a conceptually novel conditional BAC transgenic approach to unveil the distinct but interacting roles of cortical and striatal mHTT in disease pathogenesis in a human genomic transgenic mouse model of HD.
We showed that highly selective reduction of mHTT in striatal MSNs contribute significantly to the cell-autonomous amelioration of striatal postsynaptic deficits in BR mice. However, these mice still exhibit significant behavioral deficits and selective forebrain atrophy. Such findings should be interpreted with caution since our power analyses showed that much larger numbers of mice are needed to fully exclude small phenotypic improvement between BR and BACHD mice (Supplementary Table 1). Together, our results suggest that mHTT in striatal MSNs contributes to some aspects of striatal pathogenesis, but the pathogenesis of many behavioral and neurodegenerative phenotypes in BACHD likely require mHTT expression outside the striatal MSNs.
Our study is the first to evaluate the contribution of mHTT in cortical neurons in disease pathogenesis in a fl-mHTT transgenic mouse model of HD. An important finding in our study is that mHTT reduction in the Emx1-Cre lineage, relatively restricted to the CPNs, leads to partial amelioration of motor deficits and significant improvement of psychiatric-like behavioral deficits. Our study also suggests cortical mHTT genetic reduction can elicit cell-autonomous improvement of presynaptic marker protein expression, and non-cell-autonomous improvement of postsynaptic marker protein expression and synaptic neurotransmission within MSNs. This finding is consistent with our prior study demonstrating that pathological cell-cell interactions are necessary for disease pathogenesis in a mHTT fragment mouse model15. Our current study represents a significant advance by providing compelling evidence for non-cell-autonomous mHTT toxicities in striatal pathogenesis in a fl-mHTT mouse model, and by revealing that mHTT synthesized in the cortical neurons is a key source for such toxicities. Consistent with the emerging theme from other neurodegenerative disorders8, our study supports the critical roles for non-cell-autonomous disease mechanisms in the pathogenesis and possibly therapy of HD.
The most crucial finding in our current study, through the examination of BER mice, is that genetic reduction of mHTT in both cortical and striatal neurons results in the most consistent amelioration of all disease-like phenotypes in BACHD mice. Our result suggests an additive or synergistic role of cortical and striatal mHTT in HD pathogenesis. We favor the latter possibility due to the non-cell-autonomous roles for cortical mHTT in striatal synaptic pathogenesis. Certainly future studies are needed to delineate the specific, and possibly distinct, molecular pathogenic mechanisms elicited by mHTT in the cortical and striatal neurons that are underpinning the cortico-striatal pathological interactions in HD.
Finally, our study provides novel insights into where in the brain mHTT should be therapeutically targeted to ameliorate the disease. To this end, we are cognizant of some limitations of the study. First, BACHD as well as other existing HD mouse models only partially mimic the behavioral and neurodegenerative phenotypes of HD34. Our genetic experiment is also constrained by the availability of Cre mouse lines with neuronal cell-type specific expression. Moreover, the onset of Emx1-Cre and Rgs9-Cre activities is during the embryonic and perinatal ages respectively, and hence our study addresses reduction of regional mHTT synthesis in disease prevention, rather than therapeutic intervention after the disease onset. Therefore, our study cannot exclude the possibility of more significant therapeutic impacts of striatal or cortical mHTT reduction in adult ages10,11. Notwithstanding such limitations, our study provides the first proof-of-concept on the benefit of targeting mHTT in the cortical neurons to partially ameliorate multiple behavioral deficits and non-cell-autonomously improve striatal pathology in HD. More importantly, it suggests that the optimal therapeutic strategy may require the delivery of HTT-lowering therapies to both cortical and striatal neuronal targets in HD. Interestingly, strategies preferentially delivering such therapies to the cortex12 or the striatum 10,11 have been demonstrated in preclinical models. Future clinical trials will be needed to evaluate the distribution, efficacy and safety of these delivery strategies in HD patients, and to test whether the distinct but synergistic therapeutic benefits of HTT-lowering in the cortex and striatum, as predicted by our study in an HD mouse model, can also be achieved in patients.
METHODS
Generation and breeding of conditional HD mice
BACHD mice were generated and maintained in the FvB/NJ background as described3. Emx1-Cre and Rgs9-Cre mice were backcrossed into the FvB/NJ background (>10 generations) and then bred to generate Emx1-Cre/Rgs9-Cre (ER) double transgenic mice. BACHD and ER mice were bred to generate the cohort of mice with 8 different genotypes: BACHD, BE, BR, BER, Emx1-Cre, Rgs9-Cre, Emx1-Cre/Rgs9-Cre and WT. All mice were maintained and bred under standard conditions consistent with National Institutes of Health guidelines and approved by the University of California, Los Angeles Institutional Animal Care and Use Committees. Genomic DNA was prepared from mouse tails, and BACHD and various Cre lines were genotyped using established PCR protocols3,15,16.
Cre reporter mouse assay
Emx1/Rgs9 double Cre mice were mated with lacZ Cre reporter mice (Gt(ROSA)26Sortm1Sor/J) from the Jackson Laboratory (JAX003309). Brain sections from reporter mice crossed with Emx1-Cre, Rgs9-Cre or Emx1-Cre/Rgs9-Cre Cre mice (postnatal-day 90) were used to perform β-galactosidase histochemical assay using our established protocol33. Additionally, Emx1-Cre/Rgs9-Cre double transgenic mice were mated with Ai3 (Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J) Cre reporter mice from the Jackson Laboratory (JAX007903)20. Mice carrying Cre and YFP transgenes were perfused with 4% paraformaldehyde in 0.1M PBS at postnatal-day 90. Brains were post-fixed overnight at 4°C. Brain sections (20μm in thickness) were prepared using a Leica 1800 Cryostat (Deerfield) and stained with anti-NeuN (1:1000; mouse mAb, Millipore) followed by anti-mouse Alexa 594 based on our established protocols15.
Western blotting
Protein lysates and Western blots were performed using our established protocols3. Immunoblots were probed with 1C2 (specific for expanded polyglutamine repeat; Chemicon) and anti-α-tubulin (Sigma) at 1:3000 dilutions in 5% blocking solution3. For quantitation of Western blots, five mice per genotype were used in the study.
In situ hybridization
The human mHTT fragment within the two loxP sites was cloned into pCR2.1-TOPO Vector using TOPO TA Cloning Kit (Life Technologies) and used as the cDNA template. RNA was labeled with 35S-UTP using RNAMaxx High Yield Transcription Kit (Agilent) with T7 polymerase (Roche Applied Science, Indianapolis, IN). The radioactive in situ hybridization was performed using our established protocol35.
General experimental design
The sample sizes used for our experiments were pre-determined based on the power analyses for longitudinal behavioral and neuropathological studies in BACHD mice3,14. For our study, randomization and allocation of a given genotype is not necessary and hence was not performed. No individual data points were excluded from the statistical analyses. Sex-matched mice of appropriate genotypes were used for behavioral and neuropathological studies by investigators who were blinded to the genotypes.
Behavioral studies
Rotarod and Open-Field Exploration tests were performed for BACHD, WT, BE, BR and BER mice generated from BACHD x ER double-Cre at 2-, 6- and 12-months of age (n = 12–15 per genotype) using our established protocols3. The psychiatric-like behavioral tests were performed on the above mice at 12-months of age. The Light-Dark Exploration tests were performed as described before36. The Forced Swimming tests were performed using a standard protocol. Briefly, mice were allowed to acclimate to the experimental room at least one hour before testing. One mouse was put into a Plexiglas tank or a transparent plastic cylinder (15 cm diameter) filled with water (25 °C) to a height of 25 cm and videotaped for 6 minutes. The last 4 minutes of video was used to score for immobility, which was defined as absence of all movement except motions required for keeping the head above the water. Time spent immobile was recorded and analyzed. Open-Field testing was performed during the dark phase of the light/dark cycle, while other behavioral tests were performed during the light phase.
Neurodegenerative pathology
The analyses of forebrain weight, cerebellar weight, and unbiased stereological measurements of cortical and striatal volumes in BACHD, BE, BR BER and WT mice (N=8 per genotype) at 12-months of age were performed using our established protocols3,36.
Detection of presynaptic and postsynaptic marker proteins by immunofluorescence and quantitation of the indirect fluorescent signal intensities
Twelve-month old BACHD, BE, BR, BER and WT mice (n = 7–10 per genotype) were used for indirect immunofluorescent staining for Synaptophysin, Actn2 and PSD-95 in the striatum. Antibodies against synaptophysin (mouse Alexa Fluor 488 Conjugated mAb, Millipore), Actn2 (rabbit mAb, Epitomics) and PSD-95 (mouse mAb, UC Davis/NIH NeuroMab Facility) were used following manufacturer instructions with modification. Thin coronal brain sections (5μm) containing dorsal striatum (near bregma +1.2 mm) were prepared and immunostained with antibodies against synaptophysin (1:200), Actn2 (1:500) or PSD-95 (1:500). The latter two were followed by goat-anti-rabbit or goat anti-mouse secondary antibody conjugated to Alexa Fluor 594 (1:300, Invitrogen). For each mouse, two randomly selected dorsal striatal regions (near bregma +1.2 mm) were imaged at high magnification (63x) using a Zeiss LSM510 confocal laser-scanning microscope. The indirect immunofluorescence intensities of the images were measured using Image J software (NIH). All images for different genotypes were obtained and processed under identical conditions.
Slice electrophysiological recordings
Whole-cell patch clamp recordings from MSNs were obtained using standard methods26. Cells were identified by soma size, basic membrane properties (input resistance, membrane capacitance, and time constant. The patch pipette (3–5 MΩ) contained the following solution (in mM): Cs-methanesulfonate 130, CsCl 10, NaCl 4, MgCl2 1, MgATP 5, EGTA 5, HEPES 10, GTP 0.5, phosphocreatine 10, leupeptin 0.1 (pH 7.25-7.3, osmolality, 280–290 mOsm). Spontaneous postsynaptic currents (PSCs) were recorded in standard artificial cerebrospinal fluid (ACSF) composed of the following: NaCl-130 mM, NaHCO3-26 mM, KCl-3 mM, MgCl2-2 mM, NaHPO4-1.25 mM, CaCl2-2 mM, and glucose-10 mM (pH 7.4, osmolality, 300 mOsm). After characterizing the basic membrane properties of the neuron, NMDA receptor-mediated synaptic currents were evoked by electrical stimulation of afferent cortical fibers using a metal bipolar stimulating electrode. The stimulating electrode was placed on the dorsolateral corpus callosum 150–250 μm from the recording electrode. Stimuli (0.5 ms pulse duration, 0.1–1.0 mA intensity) were delivered at intervals of approximately 30 sec. Evoked NMDA receptor-mediated EPSCs were recorded at a holding potential of +40 mVin ACSF containing 10 μM bicuculline (BIC, a GABAA receptor antagonist) and 10 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX). Recording electrodes also contained QX-314 to block Na+ channel-mediated currents.
Statistical analyses
All data are shown as the mean ± standard error of the mean (SEM). SPSS 17.0 statistics software (Chicago, IL) was used to perform all of the statistical analyses. One-way Analysis Of Variance (ANOVA) followed by LSD post hoc analysis was used. The minimum significance level was set at P < 0.05.
Supplementary Material
Acknowledgments
X.W.Y. is supported by the National Institute of Neurological Disorders and Stroke (NINDS)/National Institutes of Health (NIH) grants (R01 NS049501 and R01 NS074312), the Hereditary Disease Foundation (HDF), CHDI Foundation, Inc. Neuroscience of Brain Disorders Award from The McKnight Foundation, and the David Weil Fund to the Semel Institute at UCLA. X.W.Y. is the Carol Moss Spivak Scholar in Neuroscience from UCLA Brain Research Institute. M.S.L. is supported by NINDS grant (R01 NS41574). M.S.L. and C.C. are also supported by NIH grant P30 HD004612.
Footnotes
AUTHOR CONTRIBUTIONS.
X.W.Y. provided the conceptual framework for the study, X.W.Y., N.W. and M.G. designed the experiments and discussed the results, and X.W.Y and N.W. wrote the manuscript. N.W. performed experiments shown in Fig. 1b–f, Fig. 2a–g, Fig. 3a–d, Table 1, Supplementary Table 1, Supplementary Fig. 1–5. M.G. performed experiment shown in Fig. 1e., helped to set-up the cross to generate Emx1-Cre/Rgs9-Cre mice, and performed many of the original behavioral and neuropathological studies evaluating the phenotypes of BE mice. X.L. contributed to Fig. 2h, i. E.G. and D.S. contributed to Fig. 1e. X.G. and J.C. contributed to the generation of Emx1-Cre/Rgs9-Cre mice. C.C., S.H. and M.S.L. performed experiments shown in Fig. 3e,f and Supplementary Fig. 6. H.D. helped in neuroanatomical and image analyses. Y.L. contributed to the use of Rgs9-Cre mice in our study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 5.Tabrizi SJ, et al. Potential endpoints for clinical trials in premanifest and early Huntington’s disease in the TRACK-HD study: analysis of 24 month observational data. Lancet Neurol. 2012;11:42–53. doi: 10.1016/S1474-4422(11)70263-0. [DOI] [PubMed] [Google Scholar]
- 6.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;11:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirasaki DI, et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron. 2012;75:41–57. doi: 10.1016/j.neuron.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadzi TC, et al. Assessment of cortical and striatal involvement in 523 Huntington disease brains. Neurology. 2012;79:1708–1715. doi: 10.1212/WNL.0b013e31826e9a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper SQ, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiFiglia M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menalled L, et al. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiol Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu X, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 16.Dang MT, et al. Disrupted motor learning and long-term synaptic plasticity in mice lacking NMDAR1 in the striatum. Proc Natl Acad Sci USA. 2006;103:15254–15259. doi: 10.1073/pnas.0601758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasato T, et al. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soriano P. Generalized lacZ expression with the Rosa26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 20.Madisen L, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt EF, et al. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149:1152–1163. doi: 10.1016/j.cell.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matamales M, et al. Striatal medium-sized spiny neurons: identification by nuclear staining and study of neuronal subpopulations in BAC transgenic mice. PLoS One. 2009;4:e4770. doi: 10.1371/journal.pone.0004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoghbi HY, Warren ST. Neurogenetics: advancing the “next-generation” of brain research. Neuron. 2010;68:165–173. doi: 10.1016/j.neuron.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymond LA, et al. Pathophysiology of Huntington’s disease: time-dependent alterations in synaptic and receptor function. Neuroscience. 2011;198:252–273. doi: 10.1016/j.neuroscience.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goto S, Hirano A. Synaptophysin expression in the striatum in Huntington’s disease. Acta Neuropathol. 1990;80:88–91. doi: 10.1007/BF00294227. [DOI] [PubMed] [Google Scholar]
- 26.Cepeda C, et al. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J Neurosci. 2003;23:961–969. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn A, et al. Mutant huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington’s disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 28.Becanovic K, et al. Transcriptional changes in Huntington disease identified using genome-wide expression profiling and cross-platform analysis. Hum Mol Genet. 2010;19:1438–1452. doi: 10.1093/hmg/ddq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyszynski M, et al. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 30.Dunah AW, Wyszynski M, Martin DM, Sheng M, Standaert DG. alpha-actinin-2 in rat striatum: localization and interaction with NMDA glutamate receptor subunits. Brain Res Mol Brain Res. 2000;79:77–87. doi: 10.1016/s0169-328x(00)00102-9. [DOI] [PubMed] [Google Scholar]
- 31.Kennedy MB. The postsynaptic density at glutamatergic synapses. Trends Neurosci. 1997;20:264–268. doi: 10.1016/s0166-2236(96)01033-8. [DOI] [PubMed] [Google Scholar]
- 32.Fan J, Cowan CM, Zhang LY, Hayden MR, Raymond LA. Interaction of postsynaptic density protein-95 with NMDA receptors influences excitotoxicity in the yeast artificial chromosome mouse model of Huntington’s disease. J Neurosci. 2009;29:10928–10938. doi: 10.1523/JNEUROSCI.2491-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 34.Crook ZR, Housman D. Huntington’s disease: can mice lead the way to treatment? Neuron. 2011;69:423–435. doi: 10.1016/j.neuron.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 36.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.