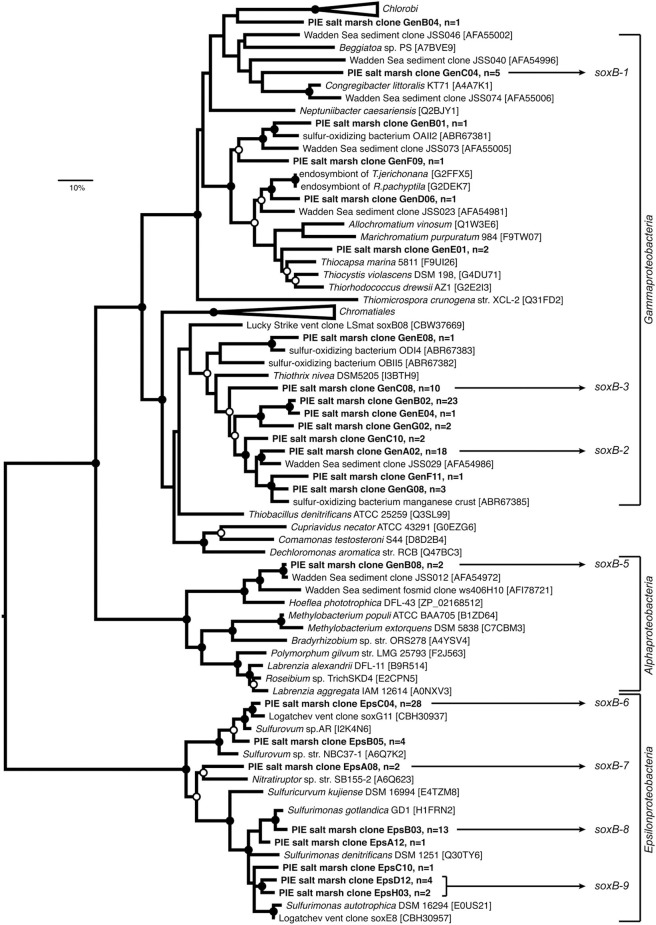
Figure 1.
Maximum-likelihood (ML) phylogenetic reconstruction of SoxB proteins deduced from sequences cloned from Plum Island Estuary salt marsh sediments (in boldface), including publicly available SoxB sequences from reference strains and uncultured bacteria (accession numbers are given). The WAG+G substitution model was used (100 re-samplings, G = 1.21, I = 0.10) based on testing different models in MEGA5. OTUs defined at 90% identity threshold are represented by one selected clone; “n” equals number of sequences per OTU. Sequences annoted as “Gen” and “Eps” were retrieved from the clone libraries prepared using the general and Epsilonproteobacteria- specific soxB primer pairs, respectively. ML bootstrap support (100 resamplings) greater than 50% (open circles) and 70% (black circles) are displayed. The tree was rooted on the Epsilonproteobacteria branch. The bar indicates 10% sequence divergence. OTUs used to design soxB primer sets targeting selected phylotypes for qPCR are shown (soxB 1–9, Table Supp1).