Figure 2.
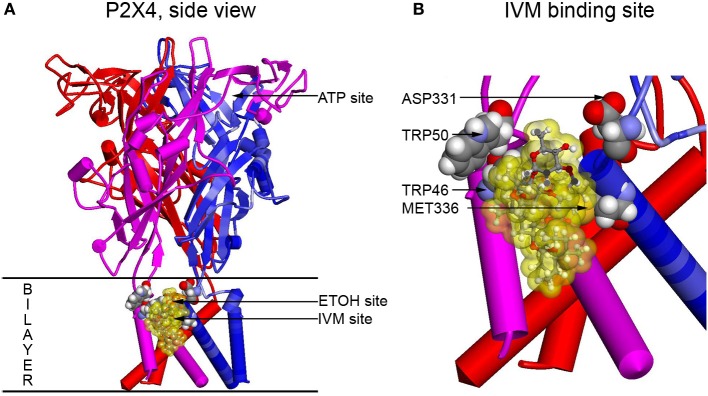
Full size and zoomed models of the rat P2X4R. A homology model of rat P2X4R was built by threading the rat primary sequence on the zebrafish X-ray structure in the open conformation (PDB ID 4DW1) (Hattori and Gouaux, 2012), essentially as described before (Popova et al., 2013). (A) A side view of the full P2X4R as viewed in the plane of the membrane looking toward the ion pore from the outside. Horizontal lines indicate the predicted extent of the membrane bilayer. The ATP binding site is over 20 Angstroms distant from the IVM binding site, yet there is substantial interaction between them. Residues important for IVM binding are rendered with space filling surfaces (carbon, oxygen, nitrogen, and hydrogen are colored gray, red, blue, and white). IVM is rendered in ball and stick and surrounded by a transparent yellow surface to outline possible interactions with the protein. A putative EtOH binding site, based on the mutations, is indicated by an arrow (EtOH site). (B) The IVM binding site is zoomed to reveal details about interactions with residues identified as important by mutagenesis. Abbreviation: P2X4R, purinergic P2X4 receptor; ATP, adenosine 5′-triphosphate; IVM, ivermectin; EtOH, ethanol.
