Figure 3.
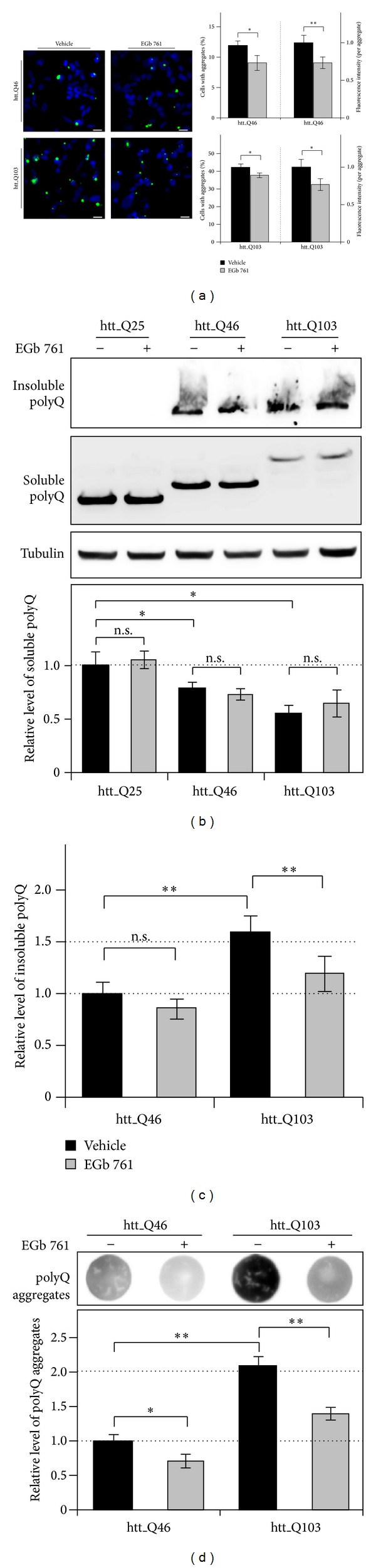
Modulating effects of EGb 761 on aggregation of polyQ proteins. (a–d) HEK293 cells expressing htt_Q46, htt_Q103, and (b) htt_Q25 were treated for 48 h with 150 μg/mL EGb 761 or vehicle. (a) Fluorescence microscopy revealed significantly more polyQ aggregates in cells expressing htt_Q103 compared to htt_Q46. Note that most aggregates are formed as nuclear inclusions. Percentage of cells exhibiting fluorescent aggregates (%) was determined by plotting total amount of polyQ aggregates against total DAPI positive cells. Total cell and aggregate amount resulted from at least three independent experiments (more than 100 cells counted). Relative fluorescence intensity (a.u.) of aggregates was quantified by calculating aggregate mean intensity with aggregate area (pixel∗au). Representative pictures are shown. Scale bar: 20 μm. (b) Cells expressing htt_Q25, htt_Q46, and htt_Q103 with EGb 761 or vehicle treatments were analyzed by immunoblotting. PolyQ proteins were detected with anti-eGFP antibody showing SDS-soluble protein bands. Cells expressing htt_Q46 and htt_Q103 showed aggregated SDS-insoluble proteins in the stacking gel (different exposure times were used). Densitometric values of soluble polyQ proteins from vehicle-treated htt_Q25-expressing cells were arbitrarily set to 1; n = 4. (c) Analysis of polyQ aggregation by using densitometric data of insoluble polyQ proteins from previous immunoblots (b). Values of vehicle-treated htt_Q46-expressing cells were arbitrarily set to 1; n = 4. (d) Whole cell extracts from cells expressing htt_Q46 and htt_Q103 with EGb 761 or vehicle treatment (samples from d) were subjected to a filter retardation assay. Detection of polyQ proteins trapped on nitrocellulose membrane revealed highly aggregated polyQ proteins in htt_Q103 compared to htt_Q46. Densitometric values of polyQ aggregates from vehicle-treated htt_Q46-expressing cells were arbitrarily set to 1; n = 4. (a–d) All values are reported as mean ± S.D. *P < 0.05 and **P < 0.01.
