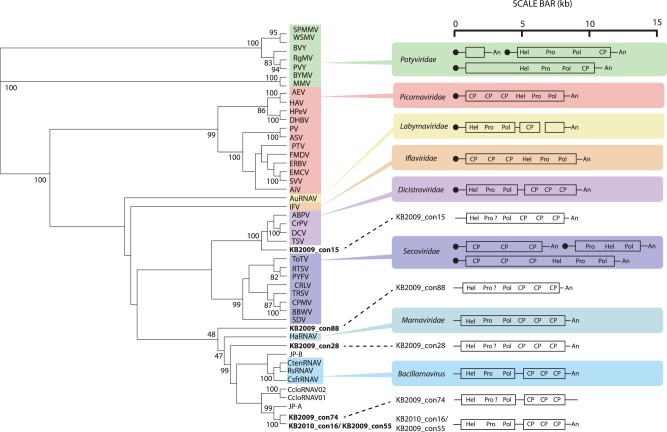
FIG 5 .
Tree of the RdRp of assembled genomes. A rooted ML tree in which the tree branches have been transformed based on an alignment of the conserved regions of the RdRP (14) from the assembled genomes from this study, homologous sequences from representative viruses from the established taxa in the order Picornavirales (highlighted in color), CcloRNAV01 and CcloRNAV02, viruses that infect a pennate diatom, and JP-A and JP-B, putative viral genomes assembled from a marine metagenome, is shown. The specific sequences in the alignment and the full names of the virus species represented by abbreviations in the figure are listed in Table S1 in the supplemental material. The statistical support values shown are percentages greater than 80 as calculated by the aLRT method. The scale bar represents 0.4 substitutions per site. Adjacent to each picornavirad taxon and environmental sequence is a schematic, based on a study by Le Gall et al. (16), that shows the gene order and genome organization for each genome type. The symbols are as follows: a black circle = “VPg” = genome-linked protein, “Hel” = helicase, “Pro” = protease, “Pol” = polymerase, “CP” = capsid protein, a black line = untranslated region, and “An” = poly(A) tail. A question mark indicates that we detected no similarity to known protease sequences but that we presume that one is encoded.