Abstract
Proteolysis carried out by different proteases control cellular processes during development and regeneration. Here we investigated the function of the proteasome and other proteases in the process of intestinal regeneration using as a model the sea cucumber Holothuria glaberrima. This echinoderm possesses the ability to regenerate its viscera after a process of evisceration. Enzymatic activity assays showed that intestinal extracts at different stages of regeneration possessed chymotrypsin-like activity. This activity was inhibited by i) MG132, a reversible inhibitor of chymotrypsin and peptidylglutamyl peptidase hydrolase (PGPH) activities of the proteasome, ii) E64d, a permeable inhibitor of cysteine proteases and iii) TPCK, a serine chymotrypsin inhibitor, but not by epoxomicin, an irreversible and potent inhibitor of all enzymatic activities of the proteasome. To elucidate the role which these proteases might play during intestinal regeneration, we carried out in vivo experiments injecting MG132, E64d and TPCK into regenerating animals. The results showed effects on the size of the regenerating intestine, cell proliferation and collagen degradation. These findings suggest that proteolysis by several proteases is important in the regulation of intestinal regeneration in H. glaberrima.
Keywords: proteasome, calpain, cathepsin, echinoderms, organogenesis
Introduction
Gastrointestinal diseases are a major problem worldwide. Each year, thousands of people are affected by diverse pathologies associated, in part, to deficiencies in the mechanisms of intestinal regeneration. Mammalian intestinal regeneration requires interaction of multiple cell types and is accomplished, among other processes, by epithelial cell migration, proliferation, apoptosis, and changes in cellular function. Several regulatory factors have been described that participate in this process (Thompson et al., 2000), however, little is known about compounds that can delay or promote intestinal regeneration. This last issue is of vital importance, since enhancement of intestinal regeneration is desirable for adequate treatment of most common intestinal pathologies.
Our laboratory has been active in studying the process of intestinal regeneration using as a model organism the sea cucumber Holothuria glaberrima. This echinoderm possesses the ability to regenerate its viscera after autotomizing and ejecting them during a process of evisceration (Hyman 1955; García-Arrarás et al., 1998). The formation of the new intestine begins by thickenings at the free end of the remaining mesenteries. During regeneration the mesenterial thickening continues increasing in size and forms the new intestinal primordium (Mashanov and García-Arrarás 2011; García-Arrarás et al., 2011). We have demonstrated that cell dedifferentiation (Murray and García-Arrarás 2004; Candelariaet al., 2006) and changes in the extracellular matrix components of the connective tissue (Quiñones et al., 2002) take place during this period. In addition, proliferation and migration of luminal epithelial cells from the cloaca and the esophagus form a lumen within the new intestine (García-Arrarás et al., 1998, 2001). As a consequence, a new intestine is formed in approximately four weeks after evisceration.
Regeneration processes have been usually regarded from the vantage point of changes in gene transcription and their translation to proteins. Nonetheless, an important part of the regulation could be mediated at the level of protein degradation. In fact, previous studies from our group showed the involvement of matrixmetalopro-teases (MMPs) in intestinal regeneration (Quiñones et al., 2002). However, this is only one class of a diverse array of proteolytic mechanisms found in cells. Other important potential sources of proteolytic activity during regenerative processes include cysteine proteases, serine proteases and proteasomes.
Several reports indicate that the proteolytic activity of cysteine proteases regulate some developmental processes. For example, the cysteine proteases cathepsin V controls cell proliferation during morphogenesis and cycling of hair follicles in mouse (Hagemannet al., 2004) and cathepsin D controls epidermal cell differentiation (Egberts et al., 2004). Caspases are essential mediators of apoptosis during development. For example, caspases control newt larval forelimb development (Vlaskalin et al., 2004) and preimplantation development in human (Spanos et al., 2002) and mouse (Exley et al., 1999). On the other hand, Ca2+ dependent proteolytic activity by calpains has also been reported during development. The inactivation of Capn4 gene, which eliminates both μ- and m-calpain activities cause embryonic death in the mouse during early organogenesis (Arthur et al., 2000). In addition, the expression of calpains 1, 2 and 3 have been shown to be regulated during early organogenesis in Xenopus (Moudilou et al., 2010).
Proteasomes, on the other hand, regulate multiple steps in the fertilization and development of marine organisms. Their participation has been demonstrated in acrosomal exocytosis triggered by the egg jelly, penetration of the vitelline coat, fusion with the egg plasma membrane and embryonic development (Sawada et al., 2002; Yokota and Sawada, 2007). Proteasomes regulate cell division during early development in mammals (Josefsberg et al., 2000; Mailhes et al., 2002) and participate during the establishment of the dorso ventral body axis in Drosophila (Klein et al., 1990) and the anterior posterior axis in C. elegans (Bowerman and Kurz, 2006). In addition, they have been showed to participate in organ formation (El-Khodor et al., 2001; Morimoto et al., 2006; Weng et al., 2006).
Serine proteases also play a role during development. In the ascidian Halocynthia, two trypsin-like proteases, spermosin and acrosin are essential for fertilization (Lambert et al., 2002). Additionally, a chymotrypsin activity has been reported to participate in fertilization (Dabrowski et al., 2004) and early development of fishes (Skern-Mauritzen et al., 2009). In chicken, trypsin-like pro-teases play a role during neuronal embryonic migration (Drapkinet al., 2002).
These antecedents suggest that specific proteolytic activities are essential for many cellular processes associated with development. In a previous report, we characterized several Ubiquitin Proteasome System (UPS) components of the sea cucumber, Holothuria glaberrima and showed that they displayed significant up-regulation during the first weeks of intestinal regeneration (Pasten et al., 2012). Moreover, we showed that when regenerating animals were treated with MG132, a proteasome inhibitor, the size of the regenerating rudiment was significantly reduced, suggesting a regulatory role of proteasomes during intestinal regeneration. The present study extends the initial investigation of the relationship between regeneration and proteolytic activities, both in vitro and in vivo. Our results confirm the important role of proteolytic activity during the intestinal regeneration process, and point to a role of calpains, and not the proteasome, as the principal mediators of proteolytic activities. In addition, they suggest that cathepsin and serine proteases might also play some roles in regulating several of the processes that take place during intestinal regeneration.
Results
Enzymatic activity during intestinal regeneration in H. glaberrima
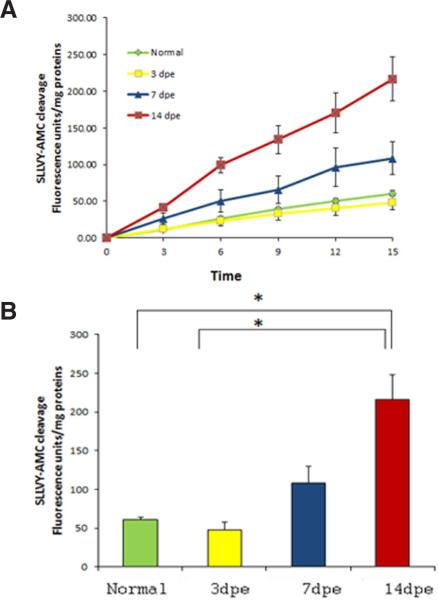
Our previous studies suggested that the proteasome could be playing an important role during intestinal regeneration in H. glaberrima (Pasten et al., 2012). Therefore, our first step was to determine if proteosomal enzymatic activity was present in extracts of regenerating intestine. To do this we used the fluorogenic substrate SLLVY-AMC which is the preferred substrate to measure the proteasome chymotrypsin-like activity, especially in marine organisms (Saitoh et al., 1993; Kawahara and Yokosawa, 1994; Tanaka et al., 2000; Yokota and Sawada, 2007). Our results showed that intestine extracts at different stages of regeneration possess enzymatic activity toward SLLVY-AMC. This activity was similar for normal and 3 day regenerating extracts, increased slightly at 7 dpe (days post evisceration) and was significantly higher at 14 dpe (Fig. 1 A,B).
Fig. 1. Enzymatic activity toward the substrate SLLVY-AMC (10μM) in crude extracts of different stages of intestinal regeneration.
Aliquots of intestinal extracts were incubated a 37°C for 15 min and the enzymatic activity was measured in a final volume of 2 ml during 15 min. (A) The cumulative cleavage of SLLVY-AMC at different time points (min) is represented. (B) Summary histogram of the results. Each bar represents the mean fluorescence units/mg proteins at 15 min of assay for each stage of intestinal regeneration ± SE. *Different from normal animals and 3dpe, p<0.05, ONE WAY ANOVA and Tukey's test.
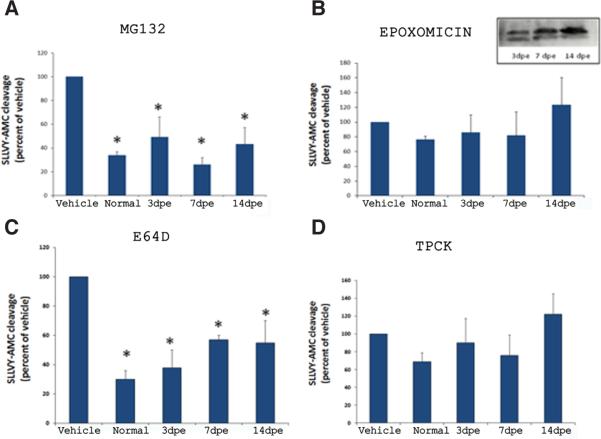
Next, we tested if this activity was inhibited by MG132, a reversible inhibitor of chymotrypsin and PGPH activity of the proteasome. The results showed that the enzymatic activities present in extracts of all regenerating intestinal stages, as well as in those from normal non-regenerating animals, were significantly inhibited (Fig. 2A). The level of inhibition was similar at all stages with extracts showing about 50% activity when compared to vehicle-treated extracts. However, since MG132 is also known to inhibit calpains and some lysosomal cathepsins (Lee and Goldberg, 1998) we decided to test the most specific proteasome inhibitor described so far, epoxomicin. This is an irreversible and potent inhibitor of the chymotrypsin-like, trypsin-like and PGPH activity of the protea-some and does not inhibit other proteases (Meng et al., 1999). In our experiments, epoxomicin did not inhibit the proteoltyic activity of the intestinal extracts (Fig. 2B). To show that proteasomes were indeed present in our homogenates, we did Western blots using the antibody that recognizes the alpha-20S proteasome subunits (Fig. 2B, insert).
Fig. 2. Effect of proteasome inhibitors on SLLVY-AMC cleavage. Intestinal extracts were prepared as described in Materials and Methods (crude extracts).
Aliquots of these extracts were incubated with MG132 10 μM (A), Epoxomicin 50 μM (B), E64d 80 μM (C) or TPCK 80 μM (D) for 15 min and the enzymatic activity was measured. Results are expressed as percent of activity compared to vehicle (100%). Each bar represents the mean fluorescence units/mg proteins at 15 min of assay for each stage ± SE. The insert in (B), represents a Western blot showing the presence of α-subunits of proteasomes in the homogenates of regenerating intestines *Different from vehicle. p<0.05, TWO WAY ANOVA and t-test. n=3.
Although SLLVY-AMC is the most widely used substrate to measure proteasomal activity, it is also commonly used to measure calpain activity (Meng et al., 1999). Therefore, the next step was to study the effect of E64d (inhibitor of calpains and some cathepsins). In addition, we also tested TPCK, an inhibitor of serine proteases. Surprisingly, the enzymatic activity was significantly inhibited by E64d in extracts from all regeneration stages and normal animals (Fig. 2C). In contrast, TPCK did not inhibit the enzymatic activity (Fig. 2D).
These results suggested that calpains and not proteasomes were responsible for the main proteolytic activity found in the extracts. However, two possibilities needed to be ruled out. First, it was possible that epoxomicin was not being effective in inhibiting holothurian proteosomal activities. To rule this out, we performed enzymatic assays using H. glaberrima gonad (sperm). We used sperm as a positive control because the sea urchin sperm contain large amounts of proteasomes and provide a rapid and sensitive method to test the effectiveness of the inhibitors (Yokota and Sawada, 2007). In our experiments, epoxomicin inhibited 90% of the proteolytic activity (Supplementary Fig. S1).
The second possibility was that the enzymatic activity of other proteases present within the extract could be masking (or interfering with) the proteasome proteolytic activity. To rule out this possibility, we employed a different protocol that provided high molecular weight (HMW) protein fractions enriched in proteasomes (Kanayama et al., 1992; Kawahara and Yokosawa 1992, 1994). The results largely confirmed the previous observations; MG132 significantly decreased the enzymatic activity present in the extracts (Supplementary Fig. S2) and a larger inhibitory effect was observed with E64d (~ 80% inhibition). However, and in contrast to the first homogenate, TPCK affected significantly the enzymatic activity in normal and at 3 dpe regenerating animals. Epoxomicin had no effect on the enzymatic activity. We also tested the effect of epoxomicin in the presence of E64d to determine if there was any residual activity due to the proteasome (Fig. 3). The results confirmed that epoxomicin had no effect on substrate hydrolysis. Thus, our experiments indicate that the activity measured by the substrate SLLVY-AMC in our intestinal extracts could not be attributed to the proteasome. These results suggest that the protease activity during intestinal regeneration in H. glaberrima is primarily due to cysteine proteases and to a lesser extent, serine proteases.
Fig. 3. Effect of epoxomicin in the presence of E64d, on the cleavage of the fluorogenic substrate SLLVY-AMC. Intestinal extracts at 7 dpe were prepared as described in methods (HMW protein fractions).
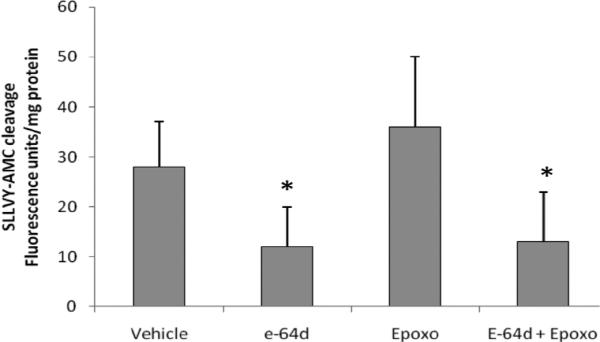
Aliquots of these extracts were incubated with E64d, epoxomicin or E64d plus epoxomicin for 15 min and the enzymatic activity to the substrate was measured for 15 min. Results are expressed as mean fluorescence units/mg protein at 15 min of assay ± SE. *p<0.05 significantly different. t-test. n=3.
In order to determine if calpains are expressed during intestinal regeneration, we studied their mRNA expression profile. Two ESTs specific for calpain 5 and 7 were obtained from a transcriptome sequencing of regenerating nerve cord of H. glaberrima. We used these sequences as templates to design specific primers (Supplementary Table S1) and to amplify the intestinal counterpart. BLAST analysis of the intestinal sequences showed high similarity to calpain 5 and 7 from a large number of species (e-value of 5e-12 for Strongylocentrotus purpuratus and e-value 3e-21 for Anolis carolinensis, calp 5 and 7 respectively). mRNA expression of these calpains was detected at all regenerations stages. Interestingly, expression of calpain 5 was significantly highest during all stages of intestinal regeneration when compared to non-regenerating animals. After 3 dpe, calpain 5 mRNA levels remained constant during the first two weeks of regeneration (Fig. 4).
Fig. 4. Gene expression profile analysis of H. glaberrima calpain-5 (A) and calpain-7 (B) during intestinal regeneration.
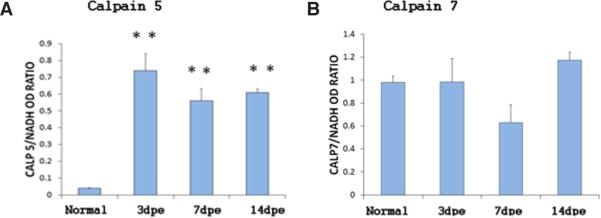
Analysis was performed by conventional RT-PCR and normalized against NADH. A significantly increase in the expression calpain 5 is observed at 3, 7 and 14 dpe compared con normal animals. Bar plot represent the mean of the normalized optical density of the bands for each regeneration stage +/− the SE. n=6 (normal, 3, and 7 dpe) and n=4 (14 dpe) for calpain 7 and n=6 for calpain 5. **Different from normal animals (N). p<0.01, ONE WAY ANOVA and Tukey's test.
In vivo experiments
To elucidate the role of these proteases during intestinal regeneration, we carried out in vivo experiments by injecting regenerating animals with MG132, E64d or TPCK. For E64d and TPCK a dose-response curve was prepared to establish 100 and 400 μM, respectively, as the appropriate concentrations for in vivo experiments (Supplementary Fig. S3).
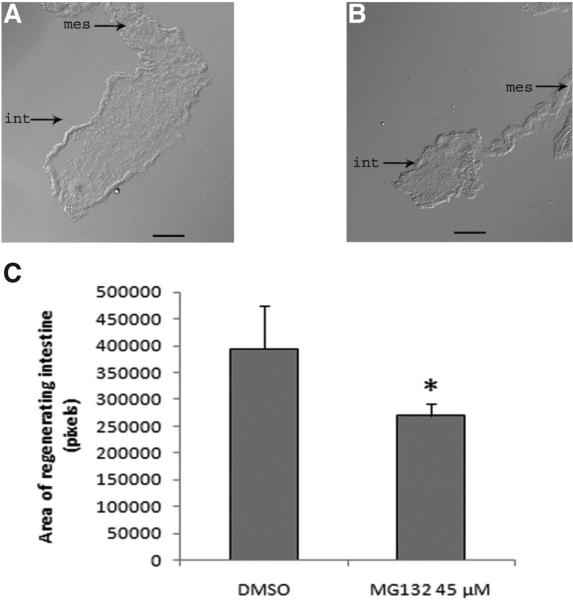
In H. glaberrima the new intestine arises from a primordium that forms from the thickening of the mesentery. The size of this structure serves as an indicator of the ongoing regenerative process. Animals treated with MG132 had a smaller regenerating structure when compared with control animals (Fig. 5 A,B). Although not statistically significant, animals treated with E64d showed a tendency toward a decrease in size of the regenerating intestine when compared with controls. TPCK did not show any significant difference in the size (data not shown).
Fig. 5. Intracoelomic injection of MG132 delays intestinal regeneration.
(A,B) Representative histological sections of DMSO-treated (control) animals (A) and MG132-treated animals (B) showing a significant reduction in the size of the regenerating intestinal primordium. (C) Measurement of the primordium area in animals treated with DMSO and MG132 shows a decrease in the regenerating intestine size of MG132-treated animals when compared with control animals (*p<0.05 t-test). Results represent the mean ± SE of at least ten animals. Scale bar, 100 μm.
The decrease in size of the regenerating intestine could be due to an effect on various cellular events that underlie the regeneration process. Among these are (1) cell proliferation (García-Arrarás et al., 1998; 2001; 2011), (2) extracellular matrix (ECM) remodeling (collagen degradation) (Quiñones et al., 2002), (3) dedifferentiation of the muscle cells (Murray and García-Arrarás 2004; Candelaria et al., 2006; García-Arrarás et al., 2011) and (4) apoptosis (Mashanov et al., 2010). We analyzed these cellular events in injected animals to determine the cellular basis of the effect of MG132, E64d and TPCK.
Cell proliferation
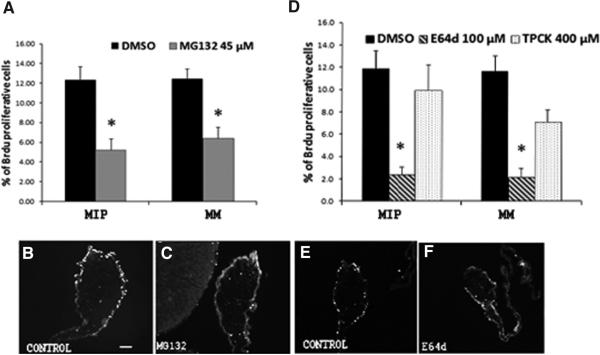
To study cell proliferation we injected BrdU for 4 hrs before animals were sacrificed. A marked effect on cell proliferation was observed in animals injected with MG132 and E64d (Fig. 6 A and D). MG132-injected animals showed a significant decrease in the percentage of BrdU labeled cells both in the mesothelium of the intestinal primordium (MIP) and of the mesenteries (MM). In control animals the percent of labeled cells was 12 ± 1 both in MIP and in MM while in those treated with MG132 it decreased to 5.2 ± 1.1 (MIP) and 6.4 ± 1.1 (MM), (Fig. 6A). A more notable effect on proliferation was observed with E64d when compared with MG132 (Fig. 6 D-F). Here, the percent of BrdU-labeled cells was 12 ± 1.1 in the DMSO-vehicle controls (MIP and MM) and 2.3 ± 0.7 in MIP and 2.1 ± 0.8 in MM in E64d treated animals. No significant effect on cell proliferation was observed with TPCK.
Fig. 6. Injections of MG132 and E64d affect cell proliferation in the regenerating intestinal primordia.
The percentage of cells that incorporated BrdU in the mesothelial layer of the intestinal primordia (MIP) and mesentery (MM) was determined in controls and animals treated with the inhibitors. Fewer mesothelial cells of animals treated with MG132 (A) and E64d (D) incorporated BrdU when compared with DMSO-treated animals. The photos below each graph are representative histological sections of animal showing a minor amount of Brdu-positive cell in animals treated with MG132 (C) and E64d (F) compared with the respective control animals (B,E). Results represent the mean ± SE of at least seven animals for MG132 and six for E64d and TPCK. *Different from DMSO (control), p<0.05. (A) t-test; (B) ONE WAY ANOVA and Tukey-test. Scale bar, 100 μm.
Extracellular matrix remodeling
During intestine regeneration in H. glaberrima an excessive remodeling of the extracellular matrix (ECM) occurs. Matrix metalloproteinases (MMPs) increase significantly and degrade several component of the ECM, among them fibrous collagen, which disappear gradually from 3 dpe to 14 dpe (Quiñones et al., 2002). Thus, the amount of collagen present at 7 dpe reflects the extent of the ECM remodelling process. To study ECM remodeling we employed a specific antibody for holothurian fibrous collagen (Quiñones et al., 2002). A notable delay in collagen degradation was observed in animals treated with E64d and TPCK (Fig. 7A). In control regenerating animals, collagen was present in 12 ± 3 % of the intestinal rudiment, while in animals treated with E64d and TPCK the percent of collagen was much higher (28 ± 4.4 % and 20 ± 2.9 % respectively). MG132 had no effect on the amount of collagen present within the regenerating structure (data not shown).
Fig. 7. Effect of E64d and TPCK injections on collagen expression.
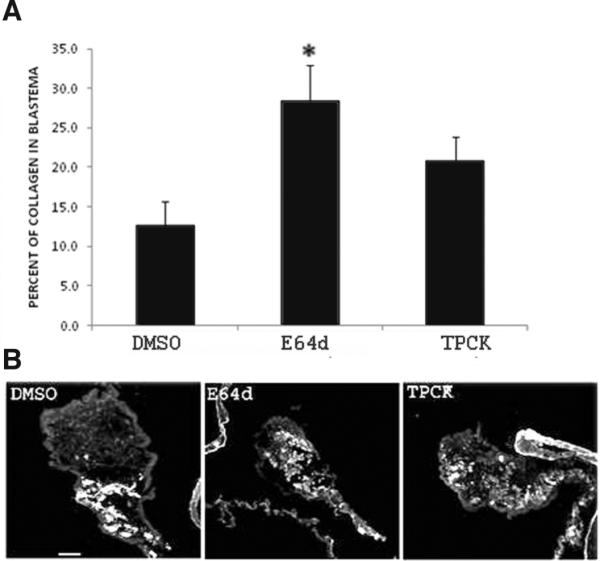
(A)Animals treated with E64d and TPCK show a delay in collagen degradation compared with DMSO (control animals). (B) Representative pictures showing the presence of collagen (white color) in animals treated with DMSO, E64d and TPCK. Results represent the mean ± SE of at least five experiments for E64d and TPCK. *Different from DMSO (control). p<0.05 (A) t-test; (B) ONE WAY ANOVA and Tukey-tes.t. Scale bar, 100 μm.
Muscle dedifferentiation
Intestinal regeneration in H. glaberrima involves the dedifferentiation of muscle cells within the mesentery (Candelaria et al., 2006, Garcia-Arraras et al., 2011). The abundance of spindle-like structures (SLSs) produced by dedifferentiating myocytes, provide an indirect measurement of the degree of ongoing dedifferentiation in the tissues (García-Arrarás and Dolmatov 2010; García-Arrarás et al., 2011). To determine if the inhibitors affected muscle dedifferentiation we studied the distribution of SLSs in regenerating intestine of controls and experimental animals. There were no significant differences in SLS distribution. Both control and experimental groups showed similar patterns of localization where the SLSs were present in the mesentery near the regenerating structure (Supplementary Fig. S4) although the primordium itself was devoid of them (data not shown).
Apoptosis
To determine if the inhibitors had an effect on cell apoptosis we employed the TUNEL assay. The percent of apoptotic cells was similar in all treatments. Although MG132 and E64d showed a trend toward an increase in the number of apoptotic cells, the effect was not statistically significant; the percent of apoptotic cells was 11.8 ± 2.8 in controls while for MG132 it was 17 ± 2.9, for E64d 18.2 ± 3 and for TPCK 8.3 ± 2.3 (Fig. 8).
Fig. 8. TUNEL assay in H. glaberrima.
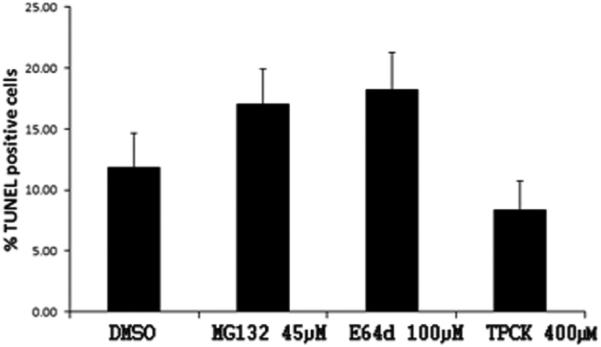
Percentage of apoptosis in mesothelial cells of the regenerating primordial of animals treated with MG132, E64d and TPCK. Results are represented as mean ± standard error. n=4 experiments. Not significantly different. ANOVA ONE WAY.
Together, these results demonstrate that the inhibition of proteolytic activities during intestinal regeneration causes some significant morphological and cellular alterations, thus suggesting that proteolytic activities play important roles in the control of intestinal regeneration.
Discussion
In this work we demonstrated that proteolytic activities are found in the regenerating intestine. We also showed that these enzymatic activities are involved in the regulation of some of the cellular processes that take part during intestinal regeneration.
Enzymatic activity assays
Our enzymatic studies focused on the activity found in extracts of normal and regenerating intestines. This activity is characterized by the ability to hydrolyze the chymotrypsin substrate SLLVY-AMC. Proteolytic activity was found in two types of preparations: crude extracts and HMW purified extracts (supplement). The activity seems to increase as the regeneration process develops with a small increase at 7 dpe and a large significant increase at 14 dpe. The increase in proteolytic activity at 14 dpe is not surprising in view that, at this stage, the intestinal luminal layer has formed (García-Arrarás et al., 1998) and luminal cells are well known to be a source of proteolytic enzymes (Klomklao, 2008).
Nature of proteolytic activity
Our initial hypothesis was that the proteasome would be responsible for the proteolytic activity within the extracts as it could be exerting a regulatory role during the process of intestinal regeneration. We had recently characterized proteasome components from H. glaberrima and shown them to be overexpressed during intestinal regeneration (Pasten et al., 2012). Thus our hypothesis was that, similar to what occurs in other systems (Glickman and Ciechanover, 2002), proteasomes could modulate different biological processes during intestinal regeneration, such as cell division or apoptosis. Results from the initial experiments using MG132 suggested that proteasomes were indeed involved in regeneration.
In our enzymatic assays, MG132 inhibited the activity to about 50% of control values at all regeneration stages studied, thus strengthening the hypothesis that the proteasome could be responsible for at least some of the observed proteolytic activity. However, MG132 is also well-known to inhibit cysteine proteases, which cleave after hydrophobic residues (similar to the chymotrypsin-like activity of the proteasomes) (Chapman, 1997). Thus, we used epoxomicin, a more selective and potent inhibitor of the proteasome that has no effect on calpain, cathepsin or any other proteases (Meng et al., 1999; Giguere and Schnellmann, 2008). We were surprised to find that epoxomicin did not inhibit the enzymatic activity in our extracts, clearly suggesting that our initial hypothesis might not be correct.
Moreover, the enzymatic activity experiments in sperm extracts clearly showed that epoxomicin could inhibit the holothurian sperm proteasomes. A possible explanation for this is that the proteolytic activity of sperm extracts is due primarily to proteasomes and that there is less (if any) activity from other proteases. In contrast, intestinal extracts might contain other proteases that concealed the activity of the proteasome. This last assumption is feasible, since the intestine is a complex system consisting of many different cell types, while the gonad content is overwhelmingly one type of highly differentiated cell. However, an effect of epoxomicin was not observed even in purified homogenates (HMW protein fractions) or when we studied the effect of epoxomicin in the presence of E64d. Thus, our present (and most logical) conclusion is that the proteolytic activity observed in our extracts does not correspond to the proteasome. This said, it is important to highlight that atypical proteasomes have been reported in invertebrate tissues. These proteasomes differ in size, subunit composition and sensitivities to protease inhibitors (Mykles, 1998). For example, differences in the composition of protein components exist between protea-some of sperm and egg in sea urchin (Saitoh, 1991) and distinct enzymatic properties have also been described for starfish oocytes proteasomes (Tanaka et al., 2000). Therefore, it is still plausible that the intestinal proteasomes of H. glaberrima are different in their sensitivity to protease inhibitors than those of the sperm and thus, not susceptible to epoxomicin inhibition. Finally, in vivo experiments with epoxomicin, which could have shown an effect of proteasome inhibition on regeneration, could not be done due to the prohibitive costs of the amount of drug needed for such studies.
Having mostly ruled out the activity of proteasomes in the extracts, our other possible candidates are calpains and cathep-sins. MG132 is also known to be an inhibitor of calpains (Lee and Goldberg 1998; Mailhes et al., 2002). Moreover, our experiments showed that E64d, a potent calpain and cathepsin inhibitor, significantly inhibited the enzymatic activity at all regeneration stages and in non-regenerating animals. In each case, the percentage of inhibition was even greater than that of MG132. Thus, the results suggest that regenerating intestines of H. glaberrima possess both calpain and cathepsin activity. Moreover, our gene expression results showed that two calpain family members, calpain 5 and calpain 7 are expressed during intestinal regeneration. Interestingly, calpain 5 remains significantly high during all process of intestinal regeneration. Additionally, in a previous report we demonstrated the presence of cathepsin C mRNA in cDNA libraries of 3- and 7-dpe regenerating intestines (Rojas-Cartagena et al., 2007). These findings are consistent with the enzymatic assays and support the presence of calpains and cathepsin during intestinal regeneration.
Calpain proteolytic activity has also been detected in different tissues of marine organisms. For example, in the muscle of the lobster (Homarus americanus), in the nerve cord of crayfish (Procambarus clarkii) and in the photoreceptor membranes in horseshoe crab (Limulus polyphermus). Endogenous substrates for these proteases are related with signal transductions and cytoeskeletal proteins (Mykles, 1998). These antecedents show that these proteases have widespread distribution and their activity could be important for regulatory processes either activating or inactivating any specific substrate or in general proteolysis.
Proteolytic nature of in vivo effects
At the organ/tissue level our results demonstrated that MG132 affected the size of the regenerating intestine. Animals injected with this inhibitor showed a smaller intestine than controls. At the cellular level, this effect cannot be attributed to changes in muscle dedifferentiation or to alterations in the ECM since our results demonstrated that these two cellular events were not altered by MG132 treatments. On the other hand the effect could be due, at least partially, to the balance between cell proliferation and cell death. MG132 caused a significant decrease in cell proliferation and a trend toward increasing apoptosis, this combination would serve to generate a smaller regenerating intestine than that found in control animals. MG132 is known to affect cell proliferation, especially in rapidly proliferating cells, such as cancer or embryonic cells. For example, MG132 affected the proliferation of lens epithelial cells (LECs) during the development of posterior capsular opacification (Awasthi and Wagner, 2006). In ovarian cancer cells, MG132 suppressed cell division in part by increasing the levels of p21 and p27 proteins (Bazzaro et al., 2006), while in gastric and colon cancer cells this inhibitor suppressed cell division via activation of BMP signaling (Wu et al., 2008).
However, MG132 was not the only inhibitor that affected cell proliferation and cell death. Our results showed that the administration of E64d also affected these processes. This inhibitor has also been shown to affect the cell cycle, suppressing cell division in Xenopus embryonic cells (Sekiguchi et al., 2002) and in sea urchin embryos, E64d inhibited cell division altering the DNA replication (Concha et al., 2005). In the regenerating intestine, although E64d effect on cell division was notably stronger than that of MG132 there was also a similar trend observed for cell death, without a measurable effect on the size of the regenerating intestine was not observed. It is possible that the reason this trend toward increased apoptosis with E64d and MG132 is not significant is due to the smaller number of animals used.
E64d also showed an effect on ECM remodeling detected as a decrease in the degradation of collagen within the intestinal primordium of animals treated with the drug. This effect was also observed with TPCK. Although we only measured the effect of the inhibitors on the presence of collagen, this itself can be used as a marker of extracellular matrix remodeling (Quiñones et al., 2002). ECM remodeling is a critical process during intestinal regeneration of H. glaberrima. Therefore, our results are in accord with other investigations in which the same inhibitors have been shown to modulate the degradation of extracellular matrix proteins (Wilsonet al., 2009).
The information obtained on the proteolytic activity of the intestinal extracts allows us to address the question of which enzymes are responsible for the in vivo effect of the inhibitors. The two main effects observed in vivo were first, an inhibition of BrdU incorporation that signaled a decline in cell proliferation and second, an increase in the presence of collagen which indicated a decrease in ECM remodeling. These effects were independent of each other in the sense that some inhibitors caused one effect (MG132-cell proliferation, TPCK -ECM remodeling) without affecting the other.
At this point, our evidence points to a role of cathepsins and serine proteases on collagen degradation. ECM collagen is subject to continuous remodeling and turnover, and its degradation involves the participation of metalloproteases, cysteine and serine proteases (Everts et al., 1995). E64d, is known to inhibit cathepsin proteolytic activities and animals injected with it showed a higher percentage of collagen within their regenerating rudiment, suggesting that collagen degradation (and thus ECM remodeling) had been inhibited. In this sense, several reports have indicated that cathepsin K, is a highly potent collagenase capable of cleaving triple helical collagens at multiple sites (Li et al., 2004). It has been demonstrated that E64d inhibits cathepsin K during lung fibrosis (Bühling et al., 2004) and in bone slices (Sondergaard et al., 2006) and that it alters the expression of MMP-9, calpain and cathepsin after transient focal cerebral ischemia in rats (Tsubokawa et al., 2006). In addition, collagen is known to be degraded within the lysosomal compartment after phagocytosis of the fibrils (Evert et al., 1996) and this intracellular digestion of phagocytosed collagen is inhibited by E64 (Everts et al., 1985). Thus, we propose that the delay on collagen degradation observed in the regenerating intestine after E64d is due to the inhibition of some cysteine pro-teases, mainly cathepsins.
In addition to cathepsins, serine proteases such as plasmin, tissue plasminogen activator, kallikreins, tryptase, chymase and elastase can also degrade ECM components (Chapman et al., 1997; Nagano et al., 2001). These proteases probably exert their function in the extracellular matrix (Cawston and Young, 2010). TPCK is a well known inhibitor of serine proteases. Therefore, the in vivo effect in collagen degradation observed after treatment with TPCK is probably due to the inhibition in the activity of some of these enzymes. However, at present we do not have sufficient information to speculate on which of the serine proteases might be the best candidate(s) to mediate this effect.
The role of extracellular enzymes on ECM remodeling had been previously studied in our laboratory (Quiñones et al., 2002). In these experiments we had shown that metalloprotease activity increased in at 7 and 14 day regenerating intestines. Subsequently, inhibition experiments injecting metalloprotease inhibitors showed a delayed in the regeneration program, which was detected by impaired early regenerative events such as a smaller size of the intestinal regenerate and reduced fibrous collagen disappearance. Here, we demonstrated that serine and cysteine proteases are also important regulators of collagen degradation during the extracellular matrix remodeling.
The effect of inhibitors on cell proliferation, on the other hand, seems to be mediated by a different group of enzymatic activity. We believe that calpains are acting as regulators of cell division during intestinal regeneration. In accord with this, MG132 and E64d, the calpain inhibitors, had a strong negative effect on cell proliferation. Consistent with this, several studies have established that calpains regulate different points of the cell cycle. Calpain inhibitors affect negatively the checkpoint G1-S, interfere with S phase and with the G2-M compartment (Jánossy et al., 2004). Cyclin D1, a positive regulator of G1 progression has been shown to be regulated posttranscriptionally by calpains (Choi et al., 1997). In addition, calpains participate in apoptosis (Sharma and Rohrer, 2004; Taylor et al., 2005). It has been demonstrated that cell cycle inhibition induces apoptosis by preventing degradation and/or processing of specific regulatory proteins (Wójcik, 2002). In our experiments, it is interesting that the same inhibitors that had an effect on cell proliferation (MG132 and E64d) showed a trend to increase the number of apoptotic cells. The fact that these differences were not statistically different might be due to the small number of animals used. Further investigations are necessary to determine the participation of these proteases in apoptosis during intestinal regeneration.
An issue that deserves special attention is the fact that several reports have emphasized a function for cysteine proteases during cell cycle. Cathepsin L isoforms devoid of a signal peptide have been shown in the nucleus of mammalian cells (Goulet et al., 2004). These have been shown to be involved in the in the control of proliferation and differentiation (Fei et al., 2007). In addition, inhibitors of cysteine proteases delay the phase S of the cell cycle (Gouletet al., 2007) and affect the embryonic cell cycle progression in the sea urchin (Morin et al., 2008). Moreover, in vivo experiments with E64d showed inhibition of sperm histones degradation which is necessary for pronuclear formation during fertilization process (Imschenetzky et al., 1997). Therefore, we cannot exclude a possible contribution of cysteine proteases (cathepsin L) on cell division during intestinal regeneration.
In summary, in this work we used protease inhibitors to study regenerating intestine proteolytic activities in vitro. Among the protease inhibitors that were most effective were MG132, a proteasome and calpain inhibitor and E64d, a calpain and cathepsin inhibitor. On the contrary, the specific proteasome inhibitor epoxomicin, did not significantly modify the activity at any stage of intestinal regeneration. In addition, we demonstrated that enzymatic activity of cysteine proteases and serine proteases are important during intestinal regeneration of H. glaberima. We propose that the enzymatic activity of calpains is necessary for the normal progression of cell cycle during the formation of the intestinal rudiment while cathepsin and serine proteases are important for collagen degradation during remodeling of extracellular matrix.
Taking together, the results provide important information on the proteolytic activities associated with intestinal regeneration of H. glaberrima. At the same time they provide insights into the cellular events where these enzymatic processes might be involved and the outcome of inhibiting their activities. Our work also opens a new perspective in the area of the medical regeneration. Since calpains are associated with different pathologies they could emerge as possible therapeutic targets.
Materials and Methods
Animals
Adult specimens of H. glaberrima were collected on the coast of Puerto Rico and kept in seawater aquaria at 20–24°C. Evisceration was induced by injecting 3–5 ml of 0.35 M KCl into the coelomic cavity (day 0) and place in aquaria.
Preparation of intestinal extracts
Regenerating (3, 7 and 14 dpe) and pieces of non-regenerating intestines (normal) were homogenized in ice-cooled 26S proteasome extraction buffer (20 mM Tris-HCl buffer, pH 8.0) containing 0.2 mM ATP, 1 mM MgCl2, 1mM DTT, 1mM EGTA, 1 % glycerol) (Kawahara and Yokosawa, 1992; 1994) and complete protease inhibitor cocktail (Calbiochem, Gibbstown, NJ). The homogenates were made with pools of two intestines (aprox. 0.2 g each). They were then centrifuged at 8000 rpm for 20 min in a table-top centrifuge at 4°C and at 51000 rpm (233800 g) g for 15 min at 4°C in an Optima L-100 XP Beckman ultracentrifuge to completely remove cell debris. The resulting supernatant was used as a crude extract for enzymatic assays. An aliquot of these fractions was used for Western blot analysis.
Preparation of proteasome-containing high molecular weight protein fractions
High molecular weights (HMW) protein fractions were prepared as described by Kanayama et al., (1992) and Kawahara and Yokosawa (1992; 1994). In these cases, a pool of four regenerating (3, 7, 14 dpe) intestines and normal intestine (aprox. 0.3 g) were homogenized in 2 ml of the proteasome extraction buffer described above. The homogenates were centrifuged at 8000 rpm for 20 min at 4°C in a table-top centrifuge and then at 50000 rpm (233800 g) for 15 min at 4°C in an Optima L-100 XP. Glycerol was added to the resulting supernatant to a final concentration of 10% and then the mixture was ultracentrifuged again at 50000 rpm (233800 g) for 5 hrs. at 4°C (Rotor Beckman type SW-50.1). The precipitate (HMW protein fractions) was dissolved in the extraction buffer and protea-some activity was assayed. Aliquots of these fractions were also used for Western blot analyses.
In other experiments, HMW protein fractions of H. glaberrima male gonad (sperm) were prepared as described above.
Enzymatic activity experiments
Unless otherwise noted, substrates and inhibitors were obtained from Enzo Life Sciences (Plymouth Meeting, PA). Chymotrypsin-like activity of the proteasome 26S was assayed at 25°C in 50 mM Tris-HCl buffer (pH 7.0) in the presence of 0.5 mM of ATP and 3.3 mM MgCl2 (Kawahara and Yokosawa 1992; 1994) using 0.02 mM of succinyl (Suc)-Leu-Leu-Val-Tyr-4-methylcoumaryl-7-amide (SLLVY-AMC) as substrate. Forty microliters of extract were added to 2 mls of buffer (described above) and incubated for 15 min at 37°C, then the substrate was added at a final concentration of 10 μM. Fluorescence was monitored with excitation at 380 nm and emission at 460 nm in Varian Cary Eclipse Fluorescence Spectophotometer (Agilent technologies, Santa Clara CA). To test the effect of the inhibitors, MG132 10 μM (chymotrypsin and PGPH proteasomal activity inhibitor), epoxomicin 50 μM (chymotrypsin, trypsin and PGPH proteasomal activity inhibitor), TPCK 80 μM (serine proteases inhibitor) and E64d 80 μM (cysteine proteases inhibitor) were used. Controls were carried out with the appropriate concentrations of the inhibitor solvent (DMSO). The protein concentration in each extract was determined using the Bradford method (Bradford, 1976).
Western blot analysis
For Western blot analyses, samples of equal protein concentration were loaded on 12-15% SDS- PAGE gel (SDS–PAGE). The samples were run for 45 min at 200 V. Proteins were transferred to PVDF membrane for 1 h at 100 V at room temperature. The membranes were blocked in 5% non-fat dry milk in TBS-T (Tris-buffered saline containing 0.1% Tween 20) for 1 hr at room temperature. The blots were then probed overnight with the antibody PW8195 (1/1000), a mouse monoclonal antibody that reacts with α1, α2, α3, α5, α6 and α7 subunits of the 20S proteasome (Enzo Life Sciences). After three washes (15 min TBS-T), the membranes were incubated with the secondary antibody (anti-mouse IgG horseradish peroxidase-linked) for 1 hr at room temperature. After three more washes, they were incubated for 5 min in super Signal West Dura Chemiluminescent Substrate (Thermo Fisher Sicentific, Rockford, IL) and the signal was visualized using a Chemi System (BioRad Molecular Imager® GelDoc™ XR).
RT-PCR experiments
ESTs for calpain 5 and 7 were obtained from a transcriptome sequencing of regenerating nerve cord of H. glaberrima. Non-eviscerated (normal) and regenerating animals at different days post-evisceration (3, 7 and 14) were anesthetized by immersion in 1,1,1-trichloro-2-methyl-2-propanol hydrate 98% or ice water for 1 h before dissection. Intestines were dissected and total RNA was extracted from intestinal homogenates using Tri-Reagent (MRC, Cincinnati, OH) and the RNeasy Mini Kit from QUIAGEN (Valencia, CA). The extracted RNA was quantified by spectrophotometry at 260-nm optical density in a NanoDrop (ND-1000) Spectrophotometer (NanoDrop Technologies, Rockland, DE). For gene-specific relative RT-PCR, first-strand synthesis was performed with the M-MLV reverse transcriptase (PROMEGA, Madison WI). In brief, 1 μg of total RNA was incubated with oligo dt for 3 min at 80°C. After incubation, dNTPs, 10× PCR Buffer RETROscript (Ambion, Austin, TX) RNase inhibitor and reverse transcriptase were added, and the reaction was incubated at 42°C for 1 hr. This was followed by 10-min incubation at 92°C. The resulting cDNA was used for PCR as described below. Primers for calpain 5 and 7 were designed for optimal performance using the Biology workbench 3.2 and Net primer free software from PREMIER Biosoft International and are shown in Supplementary Table S1. Cycling conditions for the amplified products were as follow: an initial denaturation step at 94°C for 2 min, then 34 cycles of denaturation at 94°C for 0.45 seconds, annealing at 50.8°C for 0.45 seconds and extension at 72°C for 10 min. PCR for calpain 5 was done under the same conditions, but at 36 cycles. Optical densitometric analyses of the PCR products were performed using Adobe Photoshop. Values were normalized against NADH-dehydrogenase unit 5 (NADH).
In vivo experiments
Treatments
H. glaberrima specimens were eviscerated (day 0) and placed in aquaria. Animals were injected intracoelomically with 400 μl of MG132 at a concentration of 0.9 mM, E64d at a concentration of 2 mM and TPCK at a concentration 8 mM. The volume of coelomic fluid is approximately 8 ml; therefore, the final concentration of the inhibitors in the coelom has been calculated to be approximately 45 μM for MG132, 100 μM for E64d and 400 μM for TPCK. In every case, DMSO was injected in control animals at final concentrations ranging from 0.2 to 0.5%. Each animal was injected twice a day (one injection every 12 hours) on days 5 and 6 of regeneration. On the morning of day 7, animals were injected with 50 mg/Kg of BrdU (5-bromo-2-deoxyuridine). The animals were maintained in aquaria for 4 hr and then anesthetized by immersion in a 0.2% solution of 1,1,1-trichloro-2-methyl-2-propanol hydrate 98% for 45 min and sacrificed. The intestines were then dissected and fixed in 4% paraformaldehyde at 4°C overnight. Next day they were rinsed three times for 15 min with phosphate buffer saline (PBS) and left in PBS containing 30% sucrose at 4°C for at least 24hr before embedded in Tissue-Tek. O.C.T. compound (Sakura Finetek, USA, Inc). Tissue blocks were kept at −20°C until used. Cryostat tissue sections were cut (20 μm), mounted on Poly-L-lysine-coated slides and kept at room temperature until used. A minimum of four animals was used for each inhibition experiment.
Cell and tissue measurements and analyses
Transverse sections of regenerating intestines from at least 4 animals that had received the treatments mentioned above were analyzed for several parameters of intestinal regeneration at 7 dpe. We monitored: size of the regenerating intestine, cell proliferation, collagen expression, dedifferentiation and apoptosis. A minimum of four sections (for size, dedifferentiation, apoptosis and collagen expression) and eight sections (for cell proliferation) were examined. The values corresponding to each section were averaged to obtain the value per animal.
Regenerating organ size
In animals at 7 dpe, the regenerating intestine consists of a thickening of the tip of the mesentery. This thickening forms a solid rod that extends from the esophageal region to the cloaca (García-Arrarás et al., 1998, 2011). Sections were examined and photographed using the software SpotBasic 4.7. Next, using the software ImageJ, the contours of the regenerating structure were delineated and the number of pixel was measured.
Cell proliferation
To study cell proliferation, we employed BrdU immunochemistry as described previously (Murray and García-Arrarás 2004 and Mashanov et al., 2010). Animals were treated with BrdU during 2 hrs before sacrificed. Briefly, the cryostat tissue sections of regenerating intestines were blocked with 10% goat serum. To permeabilize cell membranes, the tissue was incubated with Triton X-100 (0.15%) for 15 min, washed twice in PBS (15 min/each) and then immersed in 0.05 M HCl for 1 hour. Then, the slides were washed one more time with PBS 0.1M before incubation overnight with anti-BrdU monoclonal antibody (GE Healthcare) (1:5) in a wet chamber. The secondary antibody was GAM-ALEXA 488 (1/200). After three washes with PBS the slides were mounted in glycerol-PBS containing 4’, 6-diamidino-2-phenylindole (DAPI) to stain the cell nuclei and examined on a UV fluorescence microscope (Leitz Laborlux). Controls were done by incubating only with the secondary antibody. The percentage of BrdU-positive cells in the mesothelial layer of the mesentery (MM) next to the regenerating intestine was calculated by counting BrdU-positive cells and DAPI-positive nuclei at 40X magnification and determining the BrdU/ DAPI ratio. To determine the percent of positive cells in the mesothelium of the intestinal primordium (MIP), BrdU-positive cells and DAPI-nuclei were counted in three different areas of the primordium; two areas were in the lateral sides of the regenerating rudiment and one close to the tip. The percent of labeled cells in each area was calculated and averaged to obtain the percent of positive cells per section.
Extracellular matrix remodeling- collagen expression
Collagen expression was measured using HgfCOL, a monoclonal antibody raised against intestinal sea cucumber fibrous collagen (Quiñones et al., 2002). Fixed intestinal regenerating tissues from control and experimental animals were incubated with the antibody overnight. Then, the tissue sections were washed three times with PBS 0.1M for 15 min and placed again in the wet chamber with GAM Cy3 secondary antibody at 1/2000 (Sigma) for one hour. Later, sections were rinsed with PBS 0.1M three times for 15 min and covered with DAPI (1/50)-PBS/Glycerol mounted medium and sealed. Images were recorded and analyzed with SpotBasic 4.7 and Image J (version 6.0; Universal Imaging, Inc) software. The percentage of collagen in the primordium was calculated by measuring the total area of the structure and the area corresponding to collagen.
Cell dedifferentiation: spindle like structure (SLS)
Previous work from our laboratory has shown that muscle cells of the remaining mesenteries can dedifferentiate forming spindle-like structures (SLS) (Candelaria et al., 2006). The localization of these structures in the regenerating intestine serves as an indicator of muscle cells undergoing dedifferentiation. The differentiation process occurs in a temporal and spatial gradient beginning at the free mesenterial edge at 1 dpe and continuing during the next two weeks. As time passes the number of SLS increases in the mesentery toward the body wall as muscle cells disappear from the areas adjacent to the regenerating structure. Supplementary Fig. S4 illustrates the classification system used to study the SLS distribution. Thus, to examine if the inhibitors affect the dedifferentiation process, the presence of SLSs was determined by labeling the contractile apparatus with FITC-Phalloidin (Sigma). FITC-Phalloidin was diluted in RIA buffer (radioimmuno assays buffer: 50 mM K2HPO4; 7.4 mM KH2PO4; 154 mM NaCl; 0.5% BSA; 1.5 mM NaN3; 1:2000) and added together with the GAM Cy3 secondary antibody. Sections were examined using a fluorescence microscope where images were recorded using the SpotBasic 4.7 software program.
Apoptosis- TUNEL reaction
Apoptosis was quantified by terminal deoxynucleotidyl transferase-mediated dUTP nicked labeling (TUNEL technique) with a Fluorescein FragEl DNA Fragmentation Detection kit (Calbiochem, Cat. QIA 39) on cryosections of the normal and regenerating intestine. The percentage of TUNEL-positive cells was calculated by counting ALEXA-positive cells and DAPI-positive nuclei at 40X magnification as was described for BrdU-cell analysis, but in micrographs obtained with 40x objective and imported into Image J processing software with the Cell Counter plugin installed.
Statistical analysis
For evaluation of statistical differences between controls and experimental groups of in vivo and enzymatic assays experiments, we employed one-way or two-ways ANOVA follows of Tukey's test or t-test for multiple comparisons. These analyses were performed using the software Excel and SpotFire. All values are reported as mean ± standard error.
Supplementary Material
Abbreviations used in this paper
- ECM
extracellular matrix
- MMP
matrix metalloproteinase
- PGPH
peptidylglutamyl peptidase hydrolase
- SLLVY-AMC
(Suc)-Leu-Leu-Val-Tyr-4-methylcoumaryl-7-amide
- SLS
spindle-like structure
Footnotes
Supplementary Material (four figures) for this paper is available at: http://dx.doi.org/10.1387/ijdb.113473cp
References
- ARTHUR JS, ELCE JS, HEGADORN C, WILLIAMS K, GREER PA. Disruption of the murine calpain smal subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol Cell Biol. 2000;20:4474–4481. doi: 10.1128/mcb.20.12.4474-4481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AWASTHI N, WAGNER BJ. Suppression of human lens epithelial cell proliferation by proteasome inhibition, a potential defense against posterior capsular opacification. Invest Ophthalmol Vis Sci. 2006;47:4482–4489. doi: 10.1167/iovs.06-0139. [DOI] [PubMed] [Google Scholar]
- BAZZARO M, LEE MK, ZOSO A, STIRLING WL, SANTILLAN A, SHIH IEM, RODEN RB. Ubiquitin proteasome system stress sensitizes ovarian cancer to proteasome inhibitor-induced apoptosis. Cancer Res. 2006;66:3754–3763. doi: 10.1158/0008-5472.CAN-05-2321. [DOI] [PubMed] [Google Scholar]
- BOWERMAN B, KURZ T. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development. 2006;133:773–784. doi: 10.1242/dev.02276. [DOI] [PubMed] [Google Scholar]
- BRADFORD M. Rapid and sensitive methods for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- BÜHLING F, RÖCKEN C, BRASCH F, HARTIG R, YASUDA Y, SAFTIG P, BRÖMME D, WELTE T. Pivotal role of cathepsin K in lung fibrosis. Am J Pathol. 2004;164:2203–2216. doi: 10.1016/S0002-9440(10)63777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANDELARIA AG, MURRAY G, FILE SK, GARCÍA-ARRARÁS JE. Contribution of mesenterial muscle dedifferentiation to intestine regeneration in the sea cucumber Holothuria glaberrima. Cell Tissue Res. 2006;325:55–65. doi: 10.1007/s00441-006-0170-z. [DOI] [PubMed] [Google Scholar]
- CAWSTON TE, YOUNG DA. Proteinases involved in matrix turnover during cartilage and bone breakdown. Cell Tissue Res. 2010;339:221–235. doi: 10.1007/s00441-009-0887-6. [DOI] [PubMed] [Google Scholar]
- CHAPMAN HA, RIESE RJ, SHI GP. Emerging roles for cysteine proteases in human biology. Annu Rev Physiol. 1997;59:63–88. doi: 10.1146/annurev.physiol.59.1.63. [DOI] [PubMed] [Google Scholar]
- CHOI YH, LEE SJ, NGUYEN P, JANG JS, LEE J, WU ML, TAKANO E, MAKI M, HENKART PA, TREPEL JB. Regulation of cyclin D1 by calpain protease. J Biol Chem. 1997;272:28479–28484. doi: 10.1074/jbc.272.45.28479. [DOI] [PubMed] [Google Scholar]
- CONCHA C, MONARDES A, EVEN Y, MORIN V, PUCHI M, IMSCHENETZKY M, GENEVIÈRE AM. Inhibition of cysteine protease activity disturbs DNA replication and prevents mitosis in the early mitotic cell cycles of sea urchin embryos. J Cell Physiol. 2005;204:693–703. doi: 10.1002/jcp.20338. [DOI] [PubMed] [Google Scholar]
- DABROWSKI K, GLOGOWSKI J, CIERESZKO A. Effects of proteinase inhibitors on fertilization in sea lamprey (Petromyzon marinus). Comp Biochem Physiol B Biochem Mol Biol. 2004;139:57–62. doi: 10.1016/j.cbpc.2004.06.010. [DOI] [PubMed] [Google Scholar]
- DRAPKIN PT, MONARD D, SILVERMAN AJ. The role of serine proteases and serine protease inhibitors in the migration of gonadotropin-releasing hormone neurons. BMC Dev Biol. 2002;2:1. doi: 10.1186/1471-213X-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EGBERTS F, HEINRICH M, JENSEN JM, WINOTO-MORBACH S, PFEIFFER S, WICKEL M, SCHUNCK M, STEUDE J, SAFTIG P, PROKSCH E, SCHÜTZE S. Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J Cell Sci. 2004;117:2295–3007. doi: 10.1242/jcs.01075. [DOI] [PubMed] [Google Scholar]
- EL-KHODOR BF, KHOLODILOV NG, YARYGINA O, BURKE RE. The expression of mRNAs for the proteasomecomplex is developmentally regulated in the rat mesencephalon. Brain Res Dev Brain Res. 2001;129:47–56. doi: 10.1016/s0165-3806(01)00181-x. [DOI] [PubMed] [Google Scholar]
- EVERTS V, BEERTSEN W, TIGCHELAAR-GUTTER W. The digestion of phagocytosed collagen is inhibited by the proteinase inhibitors leupeptin and E64. Coll Relat Re. 1985;5:315–336. doi: 10.1016/s0174-173x(85)80021-2. [DOI] [PubMed] [Google Scholar]
- EVERTS V, KORPER W, NIEHOF A, JANSEN I, BEERTSEN W. Type VI collagen is phagocytosed by fibroblasts and digested in the lysosomal apparatus: involvement of collagenase, serine proteinases and lysosomal enzymes. Matrix Biol. 1995;14:665–676. doi: 10.1016/s0945-053x(05)80030-7. [DOI] [PubMed] [Google Scholar]
- EVERTS V, VAN DER ZEE E, CREEMERS L, BEERTSEN W. Phagocytosis and intracellular digestion of collagen, its role in turnover and remodelling. Histochem J. 1996;28:229–245. doi: 10.1007/BF02409011. [DOI] [PubMed] [Google Scholar]
- EXLEY GE, TANG C, MCELHINNY AS, WARNER CM. Expression of caspase and BCL-2 apoptotic family members in mouse preimplantation embryos. Biol Reprod. 1999;61:231–239. doi: 10.1095/biolreprod61.1.231. [DOI] [PubMed] [Google Scholar]
- FEI X, QIN Z, LIANG Z. Contribution of CDP/Cux, a transcription factor, to cell cycle progression. Acta Biochim Biophys Sin (Shanghai) 2007;39:923–930. doi: 10.1111/j.1745-7270.2007.00366.x. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, DOLMATOV IY. Echinoderms: potential model systems for studies on muscle regeneration. Curr Pharm Des. 2010;16:942–955. doi: 10.2174/138161210790883426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, ESTRADA-RODGERS L, SANTIAGO R, TORRES II, DÍAZ-MIRANDA L, TORRES-AVILLÁN I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea:Echinodermata). J Exp Zool. 1998;281:288–304. doi: 10.1002/(sici)1097-010x(19980701)281:4<288::aid-jez5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, GREENBERG MJ. Visceral regeneration in holothurians. Microsc Res Tech. 2001;55:438–451. doi: 10.1002/jemt.1189. [DOI] [PubMed] [Google Scholar]
- GARCÍA-ARRARÁS JE, VALENTIN-TIRADO G, FLORES JE, ROSA RJ, RIVERA-CRUZ A, SAN MIGUEL-RUIZ JE, TOSSAS K. Cell dedifferentiation and epithelial to mesenchymal transitions during intestinal regeneration in H. glaberrima. BMC Dev Biol. 2011;11:61. doi: 10.1186/1471-213X-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIGUERE CJ, SCHNELLMANN RG. Limitations of SLLVY-AMC in calpain and proteasome measurements. Biochem Biophys Res Comm. 2008;371:578–581. doi: 10.1016/j.bbrc.2008.04.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLICKMAN M, CIECHANOVER A. The Ubiquitin-Proteasome proteolytic Pathway: destruction for the sake of construction. Physiol Rev. 2002;83:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- GOULET B, BARUCH A, MOON NS, POIRIER M, SANSREGRET LL, ERICKSON A, BOGYO M, NEPVEU A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- GOULET B, SANSREGRET L, LEDUY L, BOGYO M, WEBER E, CHAUHAN SS, NEPVEU A. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol Cancer Res. 2007;5:899–907. doi: 10.1158/1541-7786.MCR-07-0160. [DOI] [PubMed] [Google Scholar]
- HAGEMANN S, GÜNTHER T, DENNEMÄRKER J, LOHMÜLLER T, BRÖMME D, SCHÜLE R, PETERS C, REINHECKEL T. The human cysteine protease cathepsin V can compensate for murine cathepsin L in mouse epidermis and hair follicles. Eur J Cell Biol. 2004;83:775–780. doi: 10.1078/0171-9335-00404. [DOI] [PubMed] [Google Scholar]
- HYMAN L. The invertebrates: Echinodermata. McGraw-Hill; New York: 1955. [Google Scholar]
- IMSCHENETZKY M, DIAZ F, MONTECINO M, SIERRA F, PUCHI M. Identification of a cysteine protease responsible for degradation of sperm histones during male pronucleus remodeling in sea urchins. J Cell Biochem. 1997;67:304–315. [PubMed] [Google Scholar]
- JÁNOSSY J, UBEZIO P, APÁTI A, MAGÓCSI M, TOMPA P, FRIEDRICH P. Calpain as a multi-site regulator of cell cycle. Biochem Pharmacol. 2004;67:1513–1521. doi: 10.1016/j.bcp.2003.12.021. [DOI] [PubMed] [Google Scholar]
- JOSEFSBERG LB, GALIANI D, DANTES A, AMSTERDAM A, DEKEL N. The proteasome is involved in the first metaphase-to-anaphase transition of meiosis in rat oocytes. Biol Reprod. 2000;62:1270–1277. doi: 10.1095/biolreprod62.5.1270. [DOI] [PubMed] [Google Scholar]
- KANAYAMA HO, TAMURA T, UGAI S, KAGAWA S, TANAHASHI N, YOSHIMURA T, TANAKA K, ICHIHARA A. Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms. Eur J Biochem. 1992;206:567–578. doi: 10.1111/j.1432-1033.1992.tb16961.x. [DOI] [PubMed] [Google Scholar]
- KAWAHARA H, YOKOSAWA H. Cell cycle-dependent change of proteasome distribution during embryonic development of the ascidian Halocynthia roretzi. Dev Biol. 1992;151:27–33. doi: 10.1016/0012-1606(92)90210-8. [DOI] [PubMed] [Google Scholar]
- KAWAHARA H, YOKOSAWA H. Intracellular calcium mobilization regulates the activity of 26 S proteasome during the metaphase-anaphase transition in the ascidian meiotic cell cycle. Dev Biol. 1994;166:623–633. doi: 10.1006/dbio.1994.1342. [DOI] [PubMed] [Google Scholar]
- KLOMKLAO S. Digestive proteinases from marine organisms and their applications. Songklanakarin J. Sci. Technol. 2008;30:37–46. [Google Scholar]
- KLEIN U, GERNOLD M, KLOETZEL PM. Cell-specific accumulation of Drosophila Proteasomes (MCP) during early development. JBC. 1990;111:2275–2282. doi: 10.1083/jcb.111.6.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMBERT C, SOMENO T, SAWADA H. Sperm Surface Proteases in Ascidian Fertilization. J. Exp Zool. 2002;292:88–95. doi: 10.1002/jez.1145. [DOI] [PubMed] [Google Scholar]
- LEE DH, GOLDBERG AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- LI Z, YASUDA Y, LI W, BOGYO M, KATZ N, GORDON RE, FIELDS GB, BRÖMME D. Regulation of collagenase activities of human cathepsins by glycosaminoglycans. J Biol Chem. 2004;279:5470–5479. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- MAILHES JB, HILLIARD C, LOWERY M, LONDON SN. MG132, an inhibitor of proteasomesand calpains, induced inhibition of oocyte maturation and aneuploidy in mouse oocytes. Chromosome. 2002;1:2. doi: 10.1186/1475-9268-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASHANOV VS, GARCÍA-ARRARÁS JE. Gut regeneration in holothurians: a snapshot of recent developments. Biol Bull. 2011;221:93–109. doi: 10.1086/BBLv221n1p93. [DOI] [PubMed] [Google Scholar]
- MASHANOV VS, ZUEVA OR, ROJAS-CATAGENA C, GARCÍA-ARRARÁS JE. Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Dev Biol. 2010;10:117. doi: 10.1186/1471-213X-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENG L, MOHAN R, KWOK BH, ELOFSSON M, SIN N, CREWS CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo anti-inflammatory activity. Proc. Natl. Acad. Sci. USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORIN V, SANCHEZ A, QUIÑONES K, HUIDOBRO JG, IRIBARREN C, BUSTOS P, PUCHI M, GENEVIÈRE AM, IMSCHENETZKY M. Cathepsin L inhibitor I blocks mitotic chromosomes decondensation during cleavage cell cycles of sea urchin embryos. J Cell Physiol. 2008;216:790–795. doi: 10.1002/jcp.21459. [DOI] [PubMed] [Google Scholar]
- MORIMOTO M, KISO M, SASAKI N, SAGA Y. Cooperative Mesp activity is required for normal somitogenesis along the anterior-posterior axis. Dev Biol. 2006;300:687–698. doi: 10.1016/j.ydbio.2006.08.043. [DOI] [PubMed] [Google Scholar]
- MOUDILOU EN, MOUTERFI N, EXBRAYAT JM, BRUN C. Calpains expression during Xenopus laevis development. Tissue and Cell. 2010;42:275–281. doi: 10.1016/j.tice.2010.07.001. [DOI] [PubMed] [Google Scholar]
- MURRAY G, GARCÍA-ARRARÁS JE. Myogenesis during holothurian intestinal regeneration. Cell Tissue Res. 2004;318:515–524. doi: 10.1007/s00441-004-0978-3. [DOI] [PubMed] [Google Scholar]
- MYKLES DL. Intracellular proteinases of invertebrates: calcium-dependent and proteasome/ubiquitin-dependent systems. Int Rev Cytol. 1998;184:157–289. doi: 10.1016/s0074-7696(08)62181-6. [DOI] [PubMed] [Google Scholar]
- NAGANO T, HAO JL, NAKAMURA M, KUMAGAI N, ABE M, NAKAZAWA T, NISHIDA T. Stimulatory effect of pseudomonal elastase on collagen degradation by cultured keratocytes. Invest Ophthalmol Vis Sci. 2001;42:1247–1253. [PubMed] [Google Scholar]
- PASTEN C, ORTIZ-PINEDA PA, GARCÍA-ARRARÁS J. Ubiquitin-proteasome system components are up-regulated during intestinal regeneration. Genesis. 2012;50:350–365. doi: 10.1002/dvg.20803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUIÑONES JL, ROSA R, RUIZ DL, GARCÍA-ARRARÁS JE. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev Biol. 2002;250:181–197. doi: 10.1006/dbio.2002.0778. [DOI] [PubMed] [Google Scholar]
- ROJAS-CARTAGENA C, ORTÍZ-PINEDA P, RAMIREZ-GOMEZ F, SUAREZ-CASTILLO EC, MATOS-CRUZ V, RODRÍGUEZ C, ORTÍZ- ZUAZAGA H, GARCÍA-ARRARÁS JE. Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol Genomics. 2007;31:203–215. doi: 10.1152/physiolgenomics.00228.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITOH Y, KAWAHARA H, MIYAMATSU H, YOKOSAWA H. Comparative studies on proteasomes (multicatalytic proteinases) isolated from spermatozoa and eggs of sea urchins. Comp Biochem Physiol B. 1991;1:71–76. doi: 10.1016/0305-0491(91)90009-3. [DOI] [PubMed] [Google Scholar]
- SAITOH Y, SAWADA H, YOKOSAWA H. High-Molecular-Weight Protease Complexes (Proteasomes) of Sperm of the Ascidian, Halocynthia roretzi: Isolation, Characterization, and Physiological Roles in Fertilization. Dev Biol. 1993;158:238–244. doi: 10.1006/dbio.1993.1182. [DOI] [PubMed] [Google Scholar]
- SAWADA H, TAKAHASHI Y, FUJINO J, FLORES SY, YOKOSAWA H. Localization and roles in fertilization of sperm proteasomes in the ascidian Halocynthia roretzi. Mol Rep Dev. 2002;62:271–276. doi: 10.1002/mrd.10089. [DOI] [PubMed] [Google Scholar]
- SEKIGUCHI T, HOSOYAMA Y, MIYATA S. Effects of proteasome inhibitor (lactacystin) and cysteine protease inhibitor (E-64-d) on processes of mitosis in Xenopus embryonic cells. Zoolog Sci. 2002;19:1251–1255. doi: 10.2108/zsj.19.1251. [DOI] [PubMed] [Google Scholar]
- SHARMA AK, ROHRER B. Calcium-induced calpain mediates apoptosis via caspase-3 in a mouse photoreceptor cell line. J Biol Chem. 2004;279:35564–35572. doi: 10.1074/jbc.M401037200. [DOI] [PubMed] [Google Scholar]
- SKERN-MAURITZEN R, FROST P, DALVIN S, KVAMME BO, SOMMERSET I, NILSEN F. A trypsin-like protease with apparent dual function in early Lepeophtheirus salmonis (Krøyer) development. BMC Mol Biol. 2009;10:44. doi: 10.1186/1471-2199-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONDERGAARD BC, HENRIKSEN K, WULF H, OESTERGAARD S, SCHURIGT U, BRÄUER R, DANIELSEN I, CHRISTIANSEN C, QVIST P, KARSDAL MA. Relative contribution of matrix metalloprotease and cysteine protease activities to cytokine-stimulated articular cartilage degradation. Osteoarthritis Cartilage. 2006;14:738–748. doi: 10.1016/j.joca.2006.01.016. [DOI] [PubMed] [Google Scholar]
- SPANOS S, RICE S, KARAGIANNIS P, TAYLOR D, BECKER DL, WINSTON RM, HARDY K. Caspase activity and expression of cell death genes during development of human preimplantation embryos. Reproduction. 2002;124:353–363. doi: 10.1530/rep.0.1240353. [DOI] [PubMed] [Google Scholar]
- TANAKA E, TAKAGI-SAWADA M, SAWADA H. Enzymatic properties of the proteasome purified from starfish oocytes and its catalytic subunits involved in oocyte maturation. Comp Biochem Physiol C Toxicol Pharmacol. 2000;125:215–223. doi: 10.1016/s0742-8413(99)00104-8. [DOI] [PubMed] [Google Scholar]
- TAYLOR RC, ADRAIN C, MARTIN SJ. Proteases, proteasomes and apoptosis: breaking Ub is hard to do. Cell Death Differ. 2005;12:1213–1217. doi: 10.1038/sj.cdd.4401703. [DOI] [PubMed] [Google Scholar]
- THOMPSON JS, SAXENA SK, SHARP JG. Regulation of intestinal regeneration: new insights. Microsc Res Tech. 2000;51:129–137. doi: 10.1002/1097-0029(20001015)51:2<129::AID-JEMT4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- TSUBOKAWA T, SOLAROGLU I, YATSUSHIGE H, CAHILL J, YATA K, ZHANG JH. Cathepsin and calpain inhibitor E64d attenuates matrix metalloproteinase-9 activity after focal cerebral ischemia in rats. Stroke. 2006;37:1888–1894. doi: 10.1161/01.STR.0000227259.15506.24. [DOI] [PubMed] [Google Scholar]
- VLASKALIN T, WONG CJ, TSILFIDIS C. Growth and apoptosis during larval forelimb development and adult forelimb regeneration in the newt (Notophthalmus viridescens). Dev Genes Evol. 2004;214:423–431. doi: 10.1007/s00427-004-0417-1. [DOI] [PubMed] [Google Scholar]
- WENG T, CHEN Z, JIN N, GAO L, LIU L. Gene expression profiling identifies regulatory pathways involved in the late stage of rat fetal lung development. Am J Physiol Lung Cell MolPhysiol. 2006;291:1027–1037. doi: 10.1152/ajplung.00435.2005. [DOI] [PubMed] [Google Scholar]
- WILSON TJ, NANNURU KC, SINGH RK. Cathepsin G-mediated activation of pro-matrix metalloproteinase 9 at the tumor-bone interface promotes transforming growth factor-beta signaling and bone destruction. Mol Cancer Res. 2009;7:1224–1233. doi: 10.1158/1541-7786.MCR-09-0028. [DOI] [PubMed] [Google Scholar]
- WÓJCIK C. Regulation of apoptosis by the ubiquitin and proteasome pathway. J Cell Mol Med. 2002;6:25–48. doi: 10.1111/j.1582-4934.2002.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU WK, SUNG JJ, WU YC, LI ZJ, YU L, CHO CH. Bone morphogenetic protein signalling is required for the anti-mitogenic effect of the proteasome inhibitor MG-132 on colon cancer cells. Br JPharmacol. 2008;154:632–638. doi: 10.1038/bjp.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOTA N, SAWADA H. Sperm proteasomes are responsible for the acro-some reaction and sperm penetration of the vitelline envelope during fertilization of the sea urchin Pseudocentrotus depressus. Dev Biol. 2007;308:222–231. doi: 10.1016/j.ydbio.2007.05.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.