Abstract
A high-throughput screen (HTS) was performed to identify molecules specifically active against Helicobacter pylori, the causative agent of peptic ulcer and gastric carcinoma. Currently, treatment of H. pylori infection is suboptimal, with failure rates approaching 25%, despite triple therapy with two broad-spectrum antibiotics and a proton pump inhibitor or quadruple therapy with added bismuth. The HTS was performed in 384-well plates, and reduction of the metabolic indicator resazurin was used as a reporter for cell growth. Diverse molecules from commercial sources were identified as hits, and in vitro validations included measurements of MIC and time-dependent killing as well as anaerobic susceptibility testing against a panel of gut microbes. In vivo validation included testing in the mouse model of H. pylori infection. The small molecule HPi1 (3-hydrazinoquinoxaline-2-thiol) had excellent potency, with an MIC of 0.08 to 0.16 μg/ml and good selectivity for H. pylori compared to a panel of commensal bacteria. HPi1 was also effective in a mouse model of H. pylori infection, reducing colony counts to below the limit of detection after oral dosing of 25 mg/kg/day for 3 days. HPi1 is a promising lead in the search for more effective and specific H. pylori therapeutics.
INTRODUCTION
Helicobacter pylori is the causative agent of peptic ulcer and gastric carcinoma (1). According to the Centers for Disease Control and Prevention, roughly every other person carries the pathogen, and there are an estimated 500,000 cases annually of active infection in the United States (2). Initial treatment is triple therapy, consisting of a proton pump inhibitor and broad-spectrum antimicrobials (usually amoxicillin and clarithromycin), or quadruple therapy, where bismuth is added to the triple therapy. According to a recent meta-analysis of clinical trials comparing triple versus quadruple therapies, 22% of patients were not cured by these two first-line treatments (3). Also of notable concern is the high degree of resistance that develops during the course of triple therapy (4, 5). Remarkably, antibiotic resistance occurred during the course of treatment in 70.6% of treatment failures (12 out of 17 subjects) (5). Worldwide resistance of H. pylori to clarithromycin and metronidazole is also increasing, which will translate into additional treatment failure (6–9). Furthermore, these aggressive therapies often have severe side effects, resulting in poor patient compliance. Considering the total number of cases, the rate of treatment failure is unacceptably high, and H. pylori is emerging as one of the most significant drug-resistant pathogens.
The current use of multiple broad-spectrum antibiotics to treat H. pylori infections was born of necessity, as monotherapies were ineffective (10, 11). H. pylori lives in close association with the gastric epithelium, and the immune system has a limited role in this environment. This makes H. pylori different from other pathogens, where partial suppression or stasis of the infection is often sufficient to cure, since the immune system eradicates the remaining bacteria. For H. pylori, complete eradication of the pathogen is required for successful treatment. Triple therapy, first developed in 1989, resulted in an effective long-term cure (12), and since then many other empirical therapies have followed suit (13). Conceptually, there is no reason why multiple broad-spectrum agents should be required, as the infection is caused by a single pathogen, the urease breath test makes diagnosis unambiguous, and recent advances in pharmacokinetic and pharmacodynamic modeling enable the prediction of doses sufficient to suppress development of resistance (14). Now, there is an increasing appreciation for the harmful effects that antibiotics have on the commensal microbiome (15), yet the development of targeted agents against H. pylori infection has lagged behind.
The current approach to treat H. pylori infection utilizes broad-spectrum antibiotics from the existing antimicrobial armamentarium. These antibiotics act against conserved essential targets and have a fairly broad spectrum of activity. Transposon mutagenesis identified 344 essential genes in H. pylori, and approximately one-quarter of them are unique to this organism (16). The high number of unique essential genes in H. pylori is likely a consequence of the bacterium exploiting a highly specialized niche in close association with epithelial cells of the stomach (17). This provides ample opportunities to discover new antibiotics with a high degree of selectivity for the pathogen. We reasoned that it should be possible to discover compounds acting specifically against H. pylori in standard commercial compound libraries that have failed to produce any useful broad-spectrum antimicrobials. We developed a high-throughput screen to identify molecules with activity against H. pylori and prioritized compounds that lacked activity against other bacteria, including gut commensals. Here, we describe the activity and selectivity of the small molecule 3-hydrazinoquinoxaline-2-thiol (HPi1) against H. pylori.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Eleven clinical isolates of H. pylori (18) and strains ATCC 43504 and SS1 (19) were used in this study. Clinical isolates were obtained from AstraZeneca (Waltham, MA). H. pylori and Campylobacter jejuni ATCC 33291 were grown overnight in brucella broth (Thermo Fisher Scientific, Waltham, MA) with 10% fetal bovine serum (FBS; American Type Culture Collection, Manassas, VA) in vented 25-cm2 tissue culture flasks. The addition of Skirrow's (20) selective medium supplement [vancomycin (10 μg/ml), trimethoprim (5 μg/ml), and polymyxin B (2.5 IU/ml)] was optional and did not affect susceptibility testing. Flasks were incubated at 37°C and 10% CO2 overnight and diluted prior to each experiment. Bacteroides fragilis ATCC 25285, Bacteroides thetaiotaomicron ATCC 29148, Staphylococcus aureus NCTC 8325-4, Streptococcus mutans ATCC 700610, Escherichia coli ATCC 700927, Enterococcus faecalis ATCC 47077, and Clostridium perfringens ATCC 13124 were cultivated in brain heart infusion broth (Becton, Dickinson, Franklin Lakes, NJ) supplemented with 5 g/liter yeast extract (Acros Organics, Morris Plains, NJ), 15 mg/liter hemin (Sigma-Aldrich, St. Louis, MO), and 1 g/liter cysteine (Sigma-Aldrich, St. Louis, MO). Bifidobacterium longum ATCC BAA-999, Lactobacillus casei ATCC 334, and Lactobacillus reuteri ATCC 23272 were cultivated in MRS broth (Oxoid, Hampshire, United Kingdom) supplemented with 1 g/liter cysteine. Clostridium difficile CD196 was cultivated in Schaedler anaerobe medium (Oxoid, Hampshire, United Kingdom). Prior to each experiment, all strains except Helicobacter and Campylobacter species were grown overnight at 37°C in an anaerobic chamber (Coy Lab Products, Grass Lake, MI).
Antibacterial agents.
Clarithromycin, amoxicillin, and vancomycin (Sigma-Aldrich, St. Louis, MO) were dissolved as 10 g/liter stock solutions in dimethyl sulfoxide (DMSO). 3-Hydrazinoquinoxaline-2-thiol, referred to as HPi1, was dissolved as 10 g/liter stock solutions in DMSO. HPi1 and other small molecules were from the ChemBridge DIVERSet library (ChemBridge, San Diego, CA). HPi1 structure was verified using nuclear magnetic resonance, and purity was determined to be >95% with liquid chromatography-mass spectrometry. For animal studies, HPi1 was dissolved in oral gavage solutions containing final concentrations of 75% polyethylene glycol 400, 20% phosphate-buffered saline, and 5% DMSO, which were made fresh each day.
IC50 dose responses.
H. pylori strain 43504 was grown overnight in brucella broth containing Skirrow's selective medium supplement and 10% FBS. Cultures were grown in vented 25-cm2 tissue culture flasks in a 10% CO2 incubator at 37°C. Dilutions of clarithromycin, amoxicillin, and HPi1 were made in 35 μl brucella broth with 10% FBS and Skirrow's supplement using an HP D300 digital dispenser (Tecan, Morrisville, NC) in 384-well black-bottom microtiter plates (Corning Inc., Corning, NY). Antibiotics were dissolved in DMSO, and the final concentration of DMSO in each well was 0.5%. A 35-μl portion of a 1:10 dilution of an overnight culture of H. pylori was added to each well using a WellMate (Thermo Fisher Scientific, Waltham, MA) liquid dispenser. Plates were incubated for 24 h, and 20 μl of 2.25 mM resazurin was added to each well. Plates were incubated for an additional 3 h, and resazurin reduction was measured using a BioTek Synergy Mx (BioTek, Winooski, VT) plate reader according to the manufacturer's instructions. The curves for the 50% inhibitory concentration (IC50) were generated using XLFit software (IDBS, London, United Kingdom).
Susceptibility tests.
The MICs for H. pylori were determined by agar dilution. H. pylori strains were grown on brucella agar plates supplemented with 7% defibrinated sheep blood for 2 days. Colonies were gathered from the plate, and cells were diluted to approximately 106 CFU per ml in brucella broth supplemented with 10% FBS. Five microliters of this bacterial suspension was used to inoculate brucella agar plates with 7% defibrinated sheep blood containing 2-fold serial dilutions of test compounds. The plates were incubated at 37°C for 4 to 6 days in a 10% CO2 incubator. MICs were also determined by broth microdilution. Strains were grown overnight at 37°C with shaking (110 rpm) in an anaerobic or hypoxic chamber. Each culture was diluted to an approximate density of 5 × 105 CFU/ml in the appropriate medium. Drug plates were prepared by aliquoting 2-fold serial dilutions of HPi1 dissolved in DMSO into clear-bottom 96-well plates. Bacterial cultures were added to each 96-well drug plate such that the final volume was 200 μl. Plates were incubated at 37°C for 24 h, and the MIC was determined visually. Final HPi1 concentrations ranged from 0.1 to 50 μg/ml, and the MIC was determined to be the lowest concentration which resulted in no turbidity.
Time-dependent killing of H. pylori by HPi1.
A frozen stock of H. pylori (ATCC 43504) was streaked onto brucella blood agar plates and incubated for 2 days at 37°C with 10% CO2. H. pylori was subcultured into vented 25-cm2 tissue culture flasks containing 10 ml of brucella broth with 10% FBS and incubated for 24 h in a hypoxic chamber (Coy Lab Products, Grass Lake, MI) with 5% O2 and 5% CO2. The culture was diluted 1:10 in fresh medium and incubated for an additional 24 h. The optical density of the culture was adjusted to 0.02 using fresh medium, and 5 ml of the suspension was added to individual wells of 6-well microtiter plates. The microtiter plates contained various concentrations of HPi1 based on multiples of the MIC, which was determined to be 0.16 μg/ml under these conditions. The cultures were incubated at 37°C with shaking and at various time points, 50 μl samples were removed, serially diluted, and plated on brucella blood agar plates. Plates were incubated microaerobically for 4 to 6 days, and viable counts were determined by counting the colonies in the highest possible dilution for each drug and time point.
In vitro pharmacokinetic profiles of HPi1.
Ninety-six-well plate-based assays were performed as previously described (21). Solubility of HPi1 was determined in a physiologically relevant buffer (system solution buffer, pH 7.4; pION Inc., Woburn, MA) after an 18-h room temperature incubation. After filtration, quantification of soluble HPi1 was determined by UV absorbance (230 to 500 nm). Using rapid equilibrium dialysis (Thermo Scientific, Rockford, IL), human plasma protein binding was determined using a modified method established by Waters and colleagues (22). HPi1 (10 μM) was incubated in human plasma (Innovative Research Inc.; Novi, MI) for 4 h and after acetonitrile-mediated protein precipitation and filtration, quantification of unbound/free HPi1 was determined by liquid chromatography mass spectroscopy (21, 22). Liver microsome stability was determined using a modified version of a previously described method (23). Briefly, HPi1 was incubated with human or mouse liver microsomes (20 mg/ml; BD Gentest, Woburn, MA) for 0, 15, 30, 60, 120, and 240 min. After centrifugation, quantification of HPi1 in the supernatant was determined using liquid chromatography-mass spectroscopy. HPi1 half-life was determined using a least-squares fit of the multiple time points based on first-order kinetics (21, 23, 24). Permeability was determined using previously described methods (21). Quantification of HPi1 that permeated a parallel artificial membrane permeability assay (pION, Woburn, MA) was determined by UV absorbance (230 to 500 nm). All assays were performed in triplicate, and the data reported here are based on two independent runs.
Mouse model of H. pylori infection.
This model has been previously described (19, 25). Briefly, 6- to 8-week-old adult specific-pathogen-free female C57BL/6 mice were obtained from Jackson Laboratories. Five mice were housed per cage and allowed to acclimate for at least 72 h after their arrival. After acclimatization, animals were inoculated by gavage, three times over 1 week, with 0.2 ml of an H. pylori SS1 suspension (∼108 CFU/dose). The following week, animals were dosed with experimental compounds or the mouse equivalent of BMT triple therapy (6.15 mg/kg bismuth subcitrate, 50 mg/kg tetracycline, and 22.5 mg/kg metronidazole), known to eradicate the infection in humans (26). The vehicle used for HPi1 and control groups was a solution containing 75% polyethylene glycol 400, 20% phosphate-buffered saline, and 5% DMSO. Dosing was performed once a day for 3 days by oral gavage. On day 4, stomachs were harvested, weighed, and homogenized, and H. pylori SS1 burden was determined by plating dilutions on H. pylori selective medium as described above (20). The presence of H. pylori infections were confirmed in initial experiments using histopathology (Giemsa staining). Histopathology was performed by the Experimental Pathology Laboratory Service Core at Boston University School of Medicine. Statistical significance was determined using a nonparametric Kruskal-Wallis test with Dunn's multiple comparisons. Statistical analyses were performed using GraphPad Prism (La Jolla, CA), and a P value of less than 0.05 was considered statistically significant. Animal use was in compliance with the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and the Office of Laboratory Animal Welfare. The animal care and use protocol was reviewed and approved by the Institutional Animal Care and Use Committee at Boston University School of Medicine.
PCR.
Colony PCR and sequencing with H. pylori-specific primers for 16S rRNA genes were used to confirm the identity of H. pylori colonies growing on selective agar from the animal experiments. Colonies from representative plates were added to 50 μl of PCR mixtures containing the primers Hp1 (5′-TGGAGAGACTAAGCCCTCC) and Hp2 (5′-ATTACTGACGCTGATTGTGC). Cycles of melting, annealing, and extension were performed at 95°C for 30 s, 49°C for 30 s, and 72°C for 2 min using a thermocycler (Bioer, Hangzhou, China). The amplified DNA fragments were sequenced (Genewiz, South Plainfield, NJ) and compared using the BLAST database of 16S rRNA gene sequences to confirm the presence or absence of H. pylori.
RESULTS
H. pylori is a fastidious microaerophile which grows inconsistently in microtiter plates. Growth of H. pylori was optimized for 384-well microtiter plates in order to screen diverse chemical libraries for molecules which directly inhibit the pathogen. Resazurin, a metabolic indicator of viability, was used as a reporter for growth. Live cells reduced the dark blue dye resazurin into resorufin, a bright pink fluorescent compound. Measuring the increase of resorufin fluorescence allowed us to monitor the growth of H. pylori after a single day of incubation and was highly reproducible, unlike optical density measurements, which were considerably less reliable. When clarithromycin was used as a positive control to emulate a hit, the Z factor (an HTS screening window coefficient, which is an indicator of signal dynamic range and data variation) was 0.7, indicating that conditions were suitable for high-throughput screening (27). A small pilot screen of approximately 30,000 molecules from the ChemBridge diversity library yielded several hits that appeared as effective as the positive control. Dose response prioritization yielded one molecule, HPi1 (Fig. 1), with excellent potency according to resazurin dose response assays (Fig. 2). The IC50 for HPi1 was 0.24 ± 0.11 μM, comparable to the broad-spectrum antibiotics clarithromycin (0.04 ± 0.01 μM) and amoxicillin (0.08 ± 0.04 μM). To confirm the results of the resazurin assay, agar dilution MIC testing was performed against a panel of H. pylori clinical isolates. The MIC against H. pylori isolates ranged from 0.002 to 0.032 μg/ml (0.01 to 0.17 μM) in the agar dilution assay (Table 1). These tests also revealed that HPi1 was effective against the clarithromycin-resistant strains ARHp172 and ARHp246 (Table 1). The high potency against H. pylori was encouraging and suggested a specific interaction between HPi1 and its target. In comparison, nonspecific antimicrobials, such as membrane-acting detergents and DNA binding agents, typically do not yield submicrogram potency.
FIG 1.

Structure of HPi1 (3-hydrazinoquinoxaline-2-thiol).
FIG 2.
Dose responses of clarithromycin, amoxicillin, and HPi1 against H. pylori ATCC 43504 in a resazurin growth inhibition assay.
TABLE 1.
Agar dilution MIC of HPi1 for a panel of H. pylori clinical isolates
| H. pylori strain | MIC (μg/ml) |
|
|---|---|---|
| HPi1 | Clarithromycin | |
| ATCC 43504 | 0.004–0.008 | 0.016–0.032 |
| SS1 | 0.002–0.004 | 0.008–0.016 |
| ARHp18 | 0.016–0.032 | 0.008–0.016 |
| ARHp23 | 0.004–0.008 | 0.016–0.064 |
| ARHp48 | 0.004–0.016 | 0.016–0.032 |
| ARHp49 | 0.008–0.016 | 0.016 |
| ARHp136 | 0.004–0.016 | 0.032 |
| ARHp143 | 0.008–0.016 | 0.016 |
| ARHp172 | 0.004–0.016 | >0.5 |
| ARHp182 | 0.004–0.008 | 0.016–0.032 |
| ARHp192 | 0.004–0.016 | 0.008–0.016 |
| ARHp209 | 0.004–0.016 | 0.064 |
| ARHp246 | 0.008–0.032 | >0.5 |
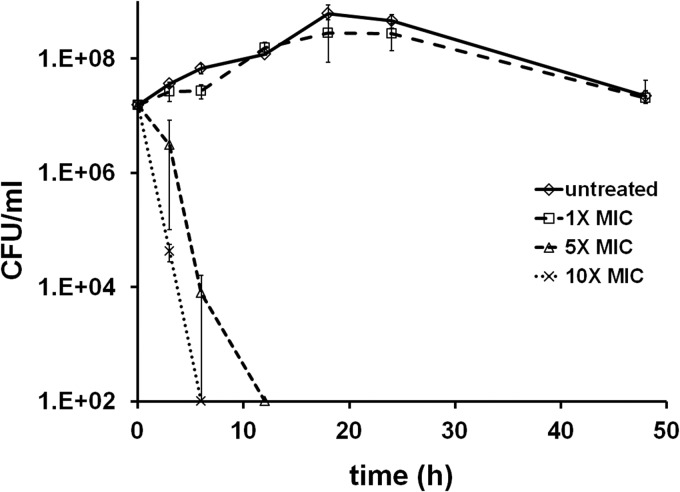
To further characterize the effect of HPi1 on H. pylori, broth dilution MIC and time-dependent killing experiments were performed. H. pylori was grown in a hypoxic chamber, and the MIC for HPi1 under these conditions was determined to be 0.08 to 0.16 μg/ml. This concentration was 20-fold higher than the agar dilution MIC, and the increased MIC may be due to an inoculum effect (28). Growing cultures of H. pylori were exposed to various concentrations of HPi1 (1-, 5-, and 10-fold the MIC). At various time points, samples were taken and plated to determine the viable cells present in each culture. A concentration of 1.6 μg/ml (10× the MIC of HPi1), was found to cause a 99% reduction in viable counts after 3 h, and no recoverable colonies were present after 6 h of exposure (Fig. 3). Similar levels of killing were caused by 5× the MIC of HPi1, although killing was delayed compared to 10× the MIC. A concentration of 0.8 μg/ml caused a 99.9% reduction in colony counts after 6 h, and 12 h of exposure was sufficient for viable counts to drop below the limit of detection (Fig. 3). No killing but a brief period of stasis was detected upon exposure of cultures to 1× the MIC of HPi1 (Fig. 3).
FIG 3.
Time-dependent killing of H. pylori by HPi1. Growing cultures of H. pylori ATCC 43504 were left untreated or exposed to HPi1 at 0.16 μg/ml, 0.8 μg/ml, and 1.6 μg/ml, which are equal to 1×, 5×, and 10× the MIC, respectively. Samples were removed, serially diluted, and plated to determine the viable counts for each starting culture and after 3, 6, 12, 18, 24, and 48 h of exposure to HPi1. The x axis is the limit of detection, and standard deviations are indicated by the error bars.
A high degree of specificity is also a desirable trait for antibiotics targeting H. pylori. To assess specificity, HPi1 was tested for activity against a panel of gut bacteria using broth microdilution MIC assays. HPi1 lacked activity against most bacteria in the panel, including the gut commensals Lactobacillus casei, Lactobacillus reuteri, and Bifidobacterium longum (Table 2). HPi1 had some activity against the Bacteroides species, but at concentrations at least 18-fold higher than the H. pylori MIC. More potent activity was detected for Campylobacter jejuni, a close relative of Helicobacter genus, with an MIC of 0.3 μg/ml.
TABLE 2.
Broth microdilution MIC of HPi1 for a panel of microaerophilic and anaerobic bacteria
| Species | Strain | HPi1 MIC (μg/ml) |
|---|---|---|
| Helicobacter pylori | ATCC 43504 | 0.08–0.16 |
| Campylobacter jejuni | ATCC 33291 | 0.3 |
| Bacteroides fragilis | ATCC 25285 | 1.5 |
| Bacteroides thetaiotaomicron | ATCC 29148 | 3.1 |
| Bifidobacterium longum | ATCC BAA-999 | >50 |
| Lactobacillus casei | ATCC 334 | >50 |
| Lactobacillus reuteri | ATCC 23272 | >50 |
| Staphylococcus aureus | NCTC 8325-4 | >50 |
| Streptococcus mutans | ATCC 700610 | >50 |
| Escherichia coli | ATCC 700927 | >50 |
| Enterococcus faecalis | ATCC 47077 | >50 |
| Clostridium perfringens | ATCC 13124 | 25 |
| Clostridium difficile | CD196 | >50 |
PubChem and ChemBank database searches revealed that HPi1 had been tested in numerous other high-throughput screens against diverse molecular and cell-based targets and was largely inactive, with no promiscuous or cytotoxic effects detected (29, 30). HPi1 was inactive in a variety of assays, including those targeting siderophore biosynthetic enzymes, DNA helicase, the CapD enzyme of B. anthracis, viral replication, actin polymerization, angiogenin RNase, the gene for the cyclin reporter Cdh1, the gene for the cyclin reporter cyclin B, E. coli FtsZ GTPase activity, eIF4G or eIF4E binding, mouse hemangioendothelioma endothelial cell proliferation, endoplasmic reticulum (ER) stress and apoptosis, herpes simplex virus (HSV) polymerase binding, lipid transfer, methyltransferase activity, Neu oncogene proliferation, proteasome inhibition, protein degradation, Pseudomonas cell wall synthesis, sulfur assimilation, tumor necrosis factor alpha (TNF-α) necrosis, and Toxoplasma invasion (29, 30).
The physicochemical and pharmacological properties of HPi1 were studied using a panel of in vitro assays to assess its potential for further advancement. This included determining the aqueous solubility (19 μg/ml), human plasma protein binding (93% bound), stability with human liver microsomes (half-life [t1/2] = 1.3 h) and the ability to passively permeate membranes using the PAMPA assay (27 × 10−6 cm/s). These are good properties, especially considering that that the compound is an unoptimized hit. The association of the pathogen with gastric epithelium may require antimicrobial action from both the lumen of the stomach and the vasculature. The ability of HPi1 to permeate membranes is a good indication that the compound will attack the pathogen from both the lumen and the tissue sites.
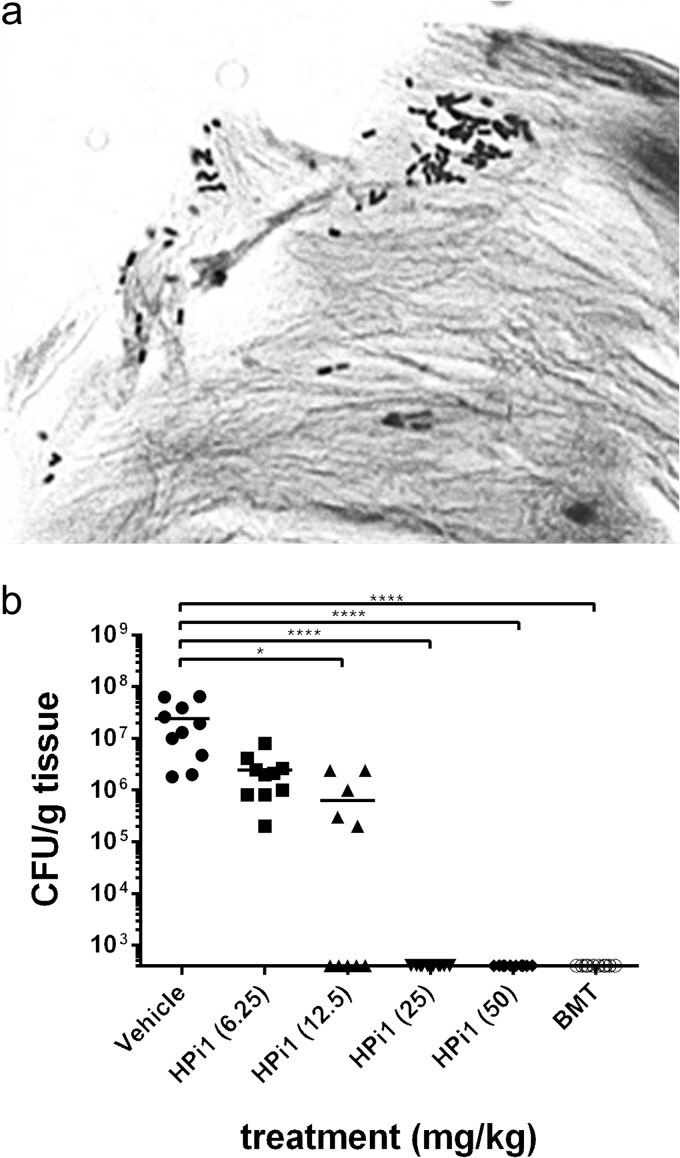
Due to the high selectivity of HPi1 against H. pylori and its good in vitro pharmacokinetic properties, we turned to a mouse model of infection to determine whether HPi1 was effective in vivo. This model uses SS1, a mouse-adapted strain, and mimics H. pylori infection in humans. Giemsa staining histopathology confirmed an abundance of H. pylori in the stomach tissue of infected mice (Fig. 4a). After the infection was established, various doses of HPi1 were delivered orally once a day for 3 days, and the effects were compared to those obtained with vehicle controls or a mouse-adapted version of triple therapy (BMT). No adverse effects were detected in mice dosed with HPi1 or BMT. One day following the last treatment, mouse stomachs were aseptically dissected, weighed, homogenized, and plated for H. pylori colony counts. HPi1 was found to decrease colony counts below the limit of detection at doses of 25 or 50 mg/kg/day, which were as effective as BMT triple therapy (Fig. 4b). Dosing with 12.5 mg/kg/day also caused significant reductions in colony counts compared to untreated controls (P < 0.05).
FIG 4.
Effect of HPi1 in a mouse model of H. pylori infection. (a) Infection with H. pylori strain SS1 was confirmed using Giemsa staining histopathology (magnification, ×1,600). (b) Each mouse was dosed by oral gavage with HPi1, BMT triple therapy, or vehicle once per day for 3 days. After treatment, stomachs were harvested, weighed, and homogenized, and H. pylori burden was determined by plating dilutions on selective medium. The x axis is the limit of detection, and horizontal bars indicate the average for each group. If no colonies were present, calculations were made using the limit of detection. *, P = 0.01 to 0.05; ****, P < 0.0001.
DISCUSSION
Novel and more effective approaches are needed to eradicate H. pylori and limit the spread of resistance. One approach is to identify chemotypes that act specifically against this pathogen without collateral damage to the beneficial host microbiota. We developed a high-throughput screen to find inhibitors of H. pylori growth. Hits were prioritized for selectivity against H. pylori by counterscreening against other bacteria present in the gastrointestinal tract. One molecule, HPi1, displayed good potency and was found to be effective in a mouse model of H. pylori infection.
There are considerable advantages for the discovery phase and for a resulting therapy identified using this selective approach. A major problem in HTS-based discovery is the very large background of promiscuous and toxic compounds, such as detergents, DNA binders, and general inhibitors of metabolism. These compounds were eliminated from further consideration by prioritizing molecules that lack activity against other bacteria, including bacteria present in the gut flora. Most known classes of antibiotics have a broad spectrum of activity, and these compounds are unlikely to be rediscovered using a selective approach. Mechanism-dependent toxicity is also unlikely for a selective antimicrobial for which the target is not present in humans. Furthermore, as a selective compound should not harm the gut microbiota, the common side effect of antibiotic-induced diarrhea may be avoided. Compound selectivity will also limit the acquisition and spread of resistance compared to broad-spectrum antibiotics.
HPi1 has a low molecular weight (192 g/mol), a good calculated octanol-water partition coefficient (0.8), and acceptable pharmaceutical properties as determined in our in vitro pharmacokinetic assays, and it displays potent in vivo efficacy. These results suggest that it is a viable lead for future medicinal chemistry efforts. PubChem (ID 781248) and ChemBank (ID 1915279) searches performed on HPi1 revealed that this compound was tested in 27 target or cell-based screens and was largely inactive. HPi1 was a primary screening hit for a single assay, i.e., replication of Vibrio cholerae chromosome II when tested at 16.6 μg/ml (31), a far higher concentration than that required to inhibit H. pylori. Taken together, these data support the high degree of selectivity that we report for HPi1. The counterscreen against gut commensals appears to be an effective way to prioritize screening hits to avoid promiscuous compounds.
Currently the target and mechanism of action of HPi1 against H. pylori are unknown. Attempts to isolate resistant mutants by serial passage on liquid or agar medium containing HPi1 were unsuccessful. Similarly, resistant mutants were not detected among colonies obtained from HPi1-treated mice. Additional resistance studies and experiments aimed at determining the mechanism of action for HPi1 are ongoing. Due to its high degree of selectivity, HPi1 may hit a target that is essential for H. pylori but absent from or not highly conserved in other bacteria. While HPi1 did cause a >4-log reduction in colony counts in the acute model of infection, an assessment of the effectiveness of monotherapy with Hpi1 in a relapsing model must await further studies, including studies assessing optimization of dosing parameters, duration of therapy, and other pharmacokinetic/pharmacodynamics properties, which are outside the scope of the current investigation.
The current state of anti-helicobacter therapy leaves room for improvement. Antibiotics with higher selectivity may lead to fewer side effects and higher patient compliance. Furthermore, new antibiotics specifically developed to eradicate H. pylori infection may yield optimized dosing regimens with improved cure rates, lower rates of resistance development, and shorter durations of therapy. The identification and characterization of HPi1 is a promising beginning toward achieving these goals.
ACKNOWLEDGMENTS
We thank the Boston University School of Medicine for access to the histopathology and animal core facilities. We thank AstraZeneca for providing the Helicobacter pylori clinical isolates.
Research reported in this publication was supported by the National Institutes of Health (R43AI098327, R44AI102452, and U54 AI057159) and by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children's Research Hospital.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Published ahead of print 31 March 2014
REFERENCES
- 1.Compare D, Rocco A, Nardone G. 2010. Risk factors in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 14:302–308 http://www.europeanreview.org/wp/wp-content/uploads/741.pdf [PubMed] [Google Scholar]
- 2.Lacy BE, Rosemore J. 2001. Helicobacter pylori: ulcers and more: the beginning of an era. J. Nutr. 131:2789S–2793S http://jn.nutrition.org/content/131/10/2789S.long [DOI] [PubMed] [Google Scholar]
- 3.Luther J, Higgins PD, Schoenfeld PS, Moayyedi P, Vakil N, Chey WD. 2010. Empiric quadruple vs. triple therapy for primary treatment of Helicobacter pylori infection: systematic review and meta-analysis of efficacy and tolerability. Am. J. Gastroenterol. 105:65–73. 10.1038/ajg.2009.508 [DOI] [PubMed] [Google Scholar]
- 4.Nishizawa T, Suzuki H, Tsugawa H, Muraoka H, Matsuzaki J, Hirata K, Ikeda F, Takahashi M, Hibi T. 2011. Enhancement of amoxicillin resistance after unsuccessful Helicobacter pylori eradication. Antimicrob. Agents Chemother. 55:3012–3014. 10.1128/AAC.00188-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilotto A, Franceschi M, Rassu M, Leandro G, Bozzola L, Furlan F, Di Mario F. 2000. Incidence of secondary Helicobacter pylori resistance to antibiotics in treatment failures after 1-week proton pump inhibitor-based triple therapies: a prospective study. Dig. Liver Dis. 32:667–672. 10.1016/S1590-8658(00)80327-8 [DOI] [PubMed] [Google Scholar]
- 6.Ogata SK, Godoy AP, da Silva Patricio FR, Kawakami E. 2013. High Helicobacter pylori resistance to metronidazole and clarithromycin in Brazilian children and adolescents. J. Pediatr. Gastroenterol. Nutr. 56:645–648. 10.1097/MPG.0b013e31828b3669 [DOI] [PubMed] [Google Scholar]
- 7.O'Connor A, Taneike I, Nami A, Fitzgerald N, Murphy P, Ryan B, O'Connor H, Qasim A, Breslin N, O'Morain C. 2010. Helicobacter pylori resistance to metronidazole and clarithromycin in Ireland. Eur. J. Gastroenterol. Hepatol. 22:1123–1127. 10.1097/MEG.0b013e328338e43d [DOI] [PubMed] [Google Scholar]
- 8.Agudo S, Alarcon T, Cibrelus L, Urruzuno P, Martinez MJ, Lopez-Brea M. 2009. High percentage of clarithromycin and metronidazole resistance in Helicobacter pylori clinical isolates obtained from Spanish children. Rev. Esp. Quimioter. 22:88–92 http://seq.es/seq/0214-3429/22/2/agudo.pdf [PubMed] [Google Scholar]
- 9.Gosciniak G, Iwanczak B, Przondo-Mordarska A, Grabinska J, Iwanczak F. 2004. High level of resistance to metronidazole and clarithromycin in Helicobacter pylori isolated from pediatric patients in Poland (1997–2001). Folia Microbiol. (Praha) 49:133–136. 10.1007/BF02931386 [DOI] [PubMed] [Google Scholar]
- 10.Glupczynski Y, Labbe M, Burette A, Delmee M, Avesani V, Bruck C. 1987. Treatment failure of ofloxacin in Campylobacter pylori infection. Lancet i:1096. [DOI] [PubMed] [Google Scholar]
- 11.McNulty CA, Gearty JC, Crump B, Davis M, Donovan IA, Melikian V, Lister DM, Wise R. 1986. Campylobacter pyloridis and associated gastritis: investigator blind, placebo controlled trial of bismuth salicylate and erythromycin ethylsuccinate. Br. Med. J. (Clin. Res. Ed.) 293:645–649. 10.1136/bmj.293.6548.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borody TJ, Cole P, Noonan S, Morgan A, Lenne J, Hyland L, Brandl S, Borody EG, George LL. 1989. Recurrence of duodenal ulcer and Campylobacter pylori infection after eradication. Med. J. Aust. 151:431–435 [DOI] [PubMed] [Google Scholar]
- 13.Chuah S-K, Tsay F-W, Hsu P-I, Wu D-C. 2011. A new look at anti-Helicobacter pylori therapy. World J. Gastroenterol. 17:3971–3975. 10.3748/wjg.v17.i35.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drusano GL. 2004. Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug'. Nat. Rev. Microbiol. 2:289–300. 10.1038/nrmicro862 [DOI] [PubMed] [Google Scholar]
- 15.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salama NR, Shepherd B, Falkow S. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926–7935. 10.1128/JB.186.23.7926-7935.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croxen MA, Ernst PB, Hoffman PS. 2007. Antisense RNA modulation of alkyl hydroperoxide reductase levels in Helicobacter pylori correlates with organic peroxide toxicity but not infectivity. J. Bacteriol. 189:3359–3368. 10.1128/JB.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carcanague D, Shue Y-K, Wuonola MA, Uria-Nickelsen M, Joubran C, Abedi JK, Jones J, Kuhler TC. 2002. Novel structures derived from 2-[[(2-pyridyl)methyl]thio]-1H-benzimidazole as anti-Helicobacter pylori agents, part 2. J. Med. Chem. 45:4300–4309. 10.1021/jm020868v [DOI] [PubMed] [Google Scholar]
- 19.Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386–1397. 10.1016/S0016-5085(97)70155-0 [DOI] [PubMed] [Google Scholar]
- 20.Skirrow MB. 1977. Campylobacter enteritis: a “new” disease. Br. Med. J. 2:9–11. 10.1136/bmj.2.6078.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.North EJ, Scherman MS, Bruhn DF, Scarborough JS, Maddox MM, Jones V, Grzegorzewicz A, Yang L, Hess T, Morisseau C, Jackson M, McNeil MR, Lee RE. 2013. Design, synthesis and anti-tuberculosis activity of 1-adamantyl-3-heteroaryl ureas with improved in vitro pharmacokinetic properties. Bioorg. Med. Chem. 21:2587–2599. 10.1016/j.bmc.2013.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters NJ, Jones R, Williams G, Sohal B. 2008. Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J. Pharm. Sci. 97:4586–4595. 10.1002/jps.21317 [DOI] [PubMed] [Google Scholar]
- 23.Di L, Kerns EH, Ma XJ, Huang Y, Carter GT. 2008. Applications of high throughput microsomal stability assay in drug discovery. Comb. Chem. High Throughput Screen. 11:469–476. 10.2174/138620708784911429 [DOI] [PubMed] [Google Scholar]
- 24.Di L, Kerns EH, Li SQ, Petusky SL. 2006. High throughput microsomal stability assay for insoluble compounds. Int. J. Pharm. 317:54–60. 10.1016/j.ijpharm.2006.03.007 [DOI] [PubMed] [Google Scholar]
- 25.Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. 1995. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science 267:1655–1658. 10.1126/science.7886456 [DOI] [PubMed] [Google Scholar]
- 26.George LL, Borody TJ, Andrews P, Devine M, Moore-Jones D, Walton M, Brandl S. 1990. Cure of duodenal ulcer after eradication of Helicobacter pylori. Med. J. Aust. 153:145–149 [DOI] [PubMed] [Google Scholar]
- 27.Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67–73. 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- 28.Berger SA, Gorea A, Moskowitz M, Santo M, Gilat T. 1993. Effect of inoculum size on antimicrobial susceptibility of Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 12:782–783. 10.1007/BF02098470 [DOI] [PubMed] [Google Scholar]
- 29.Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA. 2008. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 36:D351–D359. 10.1093/nar/gkm843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH. 2012. PubChem's BioAssay database. Nucleic Acids Res. 40:D400–D412. 10.1093/nar/gkr1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaichi Y, Duigou S, Shakhnovich EA, Waldor MK. 2009. Targeting the replication initiator of the second Vibrio chromosome: towards generation of Vibrionaceae-specific antimicrobial agents. PLoS Pathog. 5:e1000663. 10.1371/journal.ppat.1000663 [DOI] [PMC free article] [PubMed] [Google Scholar]