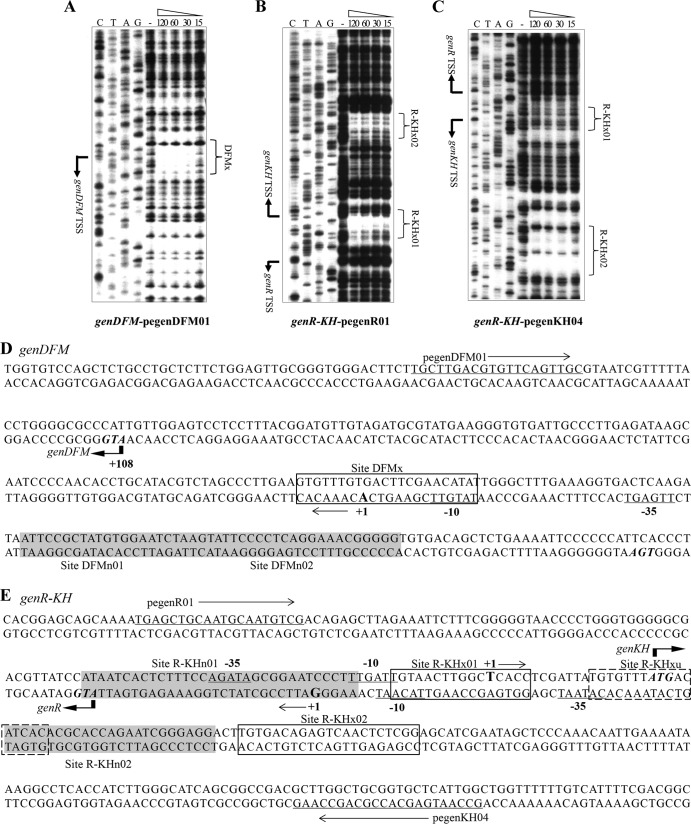
FIG 3.
Identification of the GlxR binding sites upstream of genDFM (A), genR (B), and genKH (C) operons by DNase I footprinting analysis. The reaction mixtures contained approximately 200 ng of end-labeled PCR products. These were amplified with prnagRD02 primer and 32P-labeled pegenDFM01 primer (A), pgenR01 and 32P-labeled pegenR01 (B), or pgenKH02 and 32P-labeled pegenKH04 (C) (25). Before DNase I treatment, labeled DNA was preincubated with His6-GlxR for 30 min at 25°C. Standards were generated by sequencing with 32P-labeled primers pegenDFM01(A), pegenR01 (B), and pegenKH04 (C). The concentrations (μM) of purified His6-GlxR and the GlxR-protected sequences are indicated. The nucleotide sequence around the TSS (+1) (25) is shown by solid arrows, and the GlxR binding sites are represented by brackets. Arrows indicate the direction of transcription. (D and E) Organization of the upstream region of the genDFM operon (D) and the genR-genKH promoter region (E) is shown in detail. The first codon of each gene is given in italics and boldface. The sequences in the dotted boxes are site R-KHxu as proposed previously (30), and the regulator GlxR binding sites are showed in the box. The TSSs are shown by +1 (boldface) and arrows and the putative −10 and −35 promoter sequences are underlined. The regulator GenR binding sites are highlighted in gray.