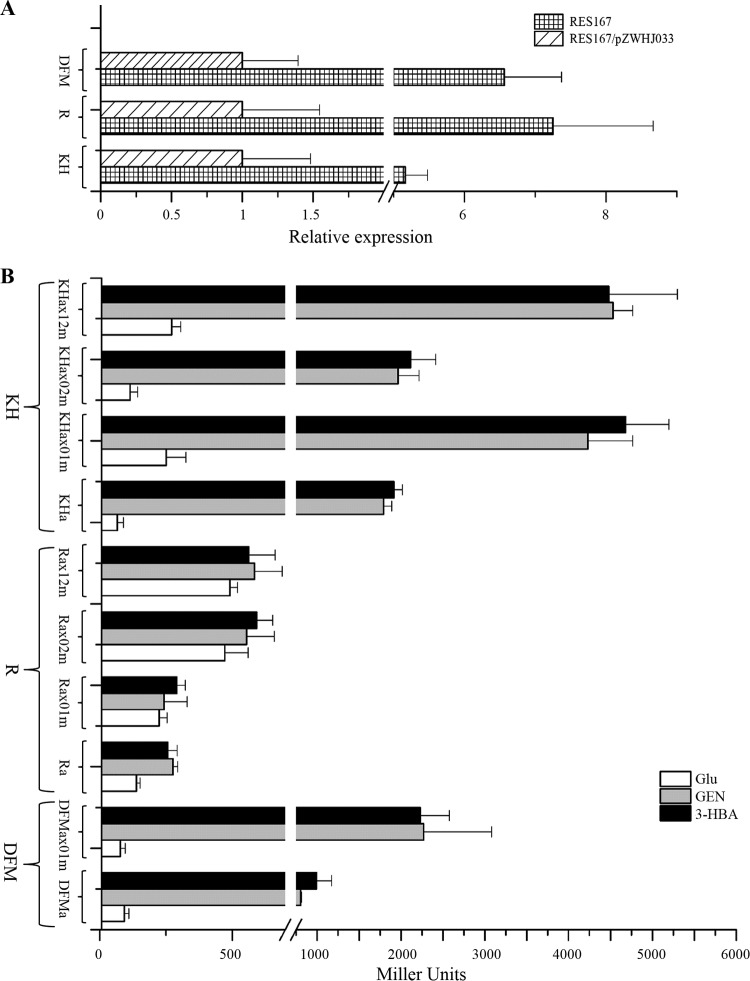
FIG 6.
GlxR attenuates upregulation of the transcription of genDFM, genR, and genKH operons. (A) qRT-PCR analyses examining the transcription of genDFM, genR, and genKH in strains RES167 and RES167/pZWHJ033. RNA samples were isolated from strains RES167 and RES167/pZWHJ033 grown on MM with 2 mM 3-HBA overnight. The levels of gene expression in each sample were calculated as the fold expression ratio after normalization to the 16S rRNA gene transcript. The values are averages of two independent RT-qPCR experiments. Error bars indicate standard deviations. These data are derived from at least three independent measurements. There was a significant difference in the transcription of genDFM, genR, and genKH between strains RES167 and RES167/pZWHJ033 (P < 0.001, paired-samples test). (B) β-Galactosidase activity driven by genDFM, genR, and genKH promoters with their corresponding GlxR binding sites in strain RES167. DFMa, Ra, and KHa containing GlxR binding sites are the GenR binding sites reported as previously (25). DFMax01m, Rax01m, Rax02m, Rax12m, KHax01m, KHax02m, and KHax12m are the constructs containing the mutated GlxR binding sites described in Materials and Methods and shown in Fig. 1. The β-galactosidase activity analyses were performed as given in the text. The data are derived from at least three independent measurements, and error bars indicate standard deviations. There was a significant difference in promoter activity between GlxR binding sites and its mutated sites from strains RES167 grown on MM with GEN or 3-HBA (P < 0.001, one-way ANOVA).