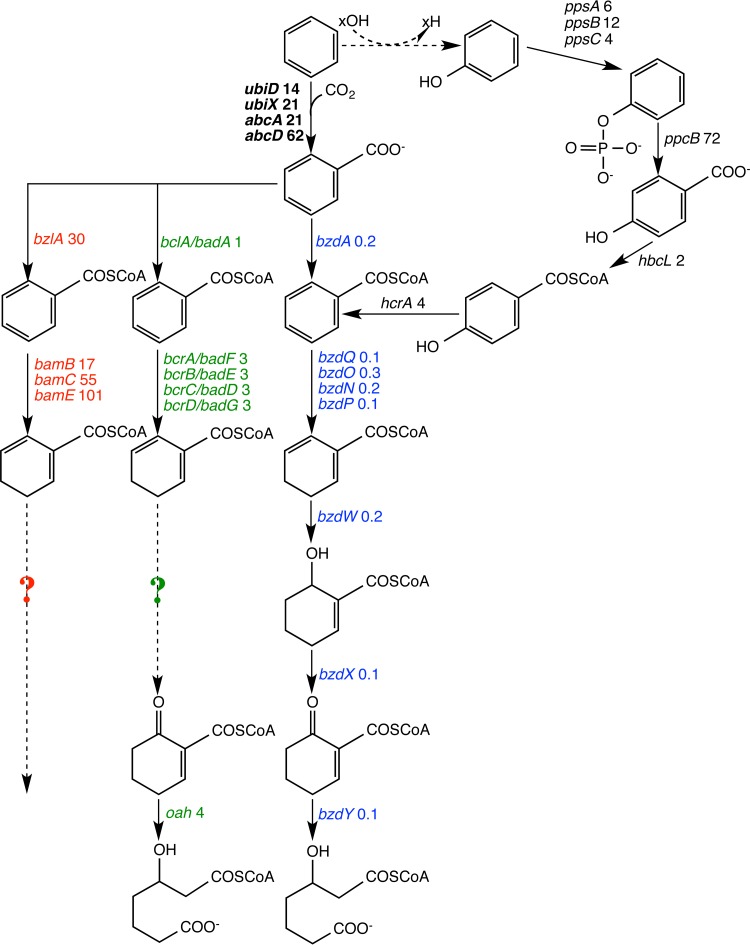
FIG 6.
Gene transcripts identified in RNA samples corresponding to known or hypothesized enzymes involved in the anaerobic metabolism of monoaromatic compounds in different anaerobes. Three parallel yet distinct pathways are known for benzoate catabolism: one found in Azoarcus/Aromatoleum spp. (bzd genes, blue), one identified in Thauera/Alphaproteobacteria spp. (bcr/bad genes, green), and the other in obligate anaerobes (bam genes, red) (54). Sequences corresponding to each of these pathways were identified in the culture. The numbers adjacent to gene names are the values for specificity (for benzene relative to benzoate). Question marks indicate where corresponding bcr/bad and bam genes were not found in the current data set. The putative carboxylase genes proposed for the Clostridium enrichment culture BF (6) are shown in boldface black, while the genes associated with the anaerobic phenol degradation pathway are shown in black but not in boldface. All contigs associated with these genes and their associated specificities are provided in Table S4 in the supplemental material.