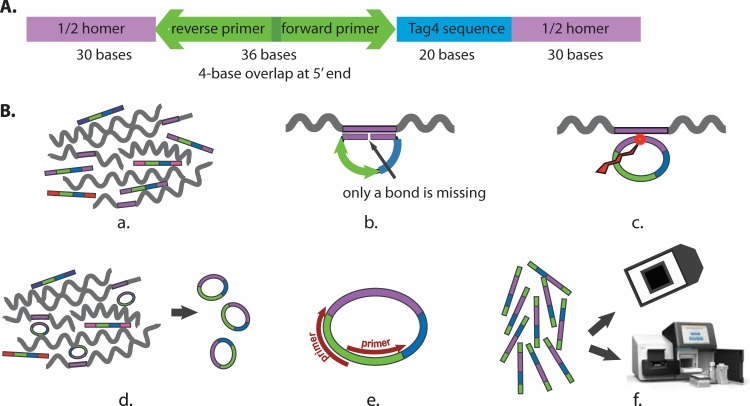
FIG 1.
Molecular probe design and assays. (A) Design of the molecular probes synthesized at the SGTC. The purple color represents the 60-base sequence homology domain (the Homer), which is divided into two 30-base segments. Blue represents the 20-base oligonucleotide barcode from the Tag4 array. Green represents the 36-base domain for the two 20-base PCR primers. The two 20-base primers overlap by 4 bases at their respective 5′ ends. The total is 116 bases. The 5′ end is phosphorylated. (B) Assays. (a) The molecular probe mixture is incubated with the denatured target DNA (wiggly lines) under annealing conditions. (b) Where sufficient sequence similarity exists between the molecular probe and the target single-stranded DNA (in purple), 60 bp of duplex DNA form. The 5′-phosphorylated end of the molecular probe is adjacent to the 3′-hydroxyl end of the probe with no bases missing. (c) Ligation creates a phosphodiester bond between the 5′ and 3′ ends. (d) Exonucleases digest the remaining single-stranded linear DNA, leaving only the single-stranded circular DNA. (e) PCR amplifies the single-stranded circular DNA. The 5′ ends of the PCR primers are biotinylated for the Tag4 readout but not for the Illumina readout. (f) The final readout is by either fluorescence on the Tag4 array (top) or sequencing by the Illumina MiSeq instrument (bottom).