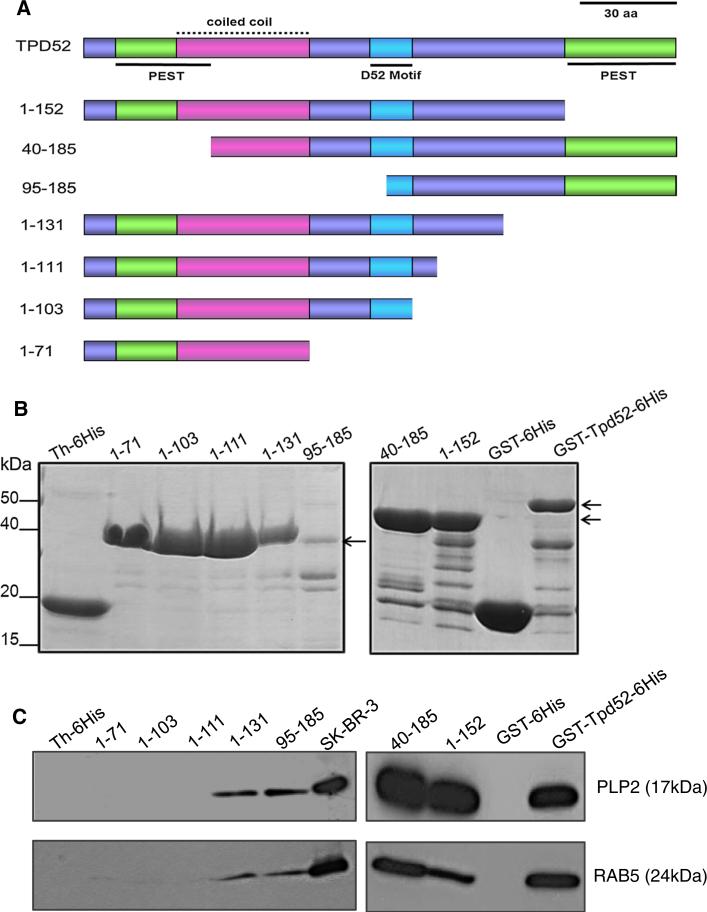
Fig. 3.
a Schematic diagram of TPD52 sequence features included in deleted TPD52/ Tpd52 proteins used in pull-down assays. PEST motifs are shown in green, underlined, the coiled-coil motif is shown in pink, with a broken line, the D52 motif is shown in blue, underlined, with all other regions shown in purple. The N-terminal PEST and coiled-coil motifs partially overlap. Amino acid co-ordinates of deleted proteins are shown at the left. Scale bar = 30 aa. b Coomassie Brilliant Blue-stained recombinant TPD52/ Tpd52 (positions highlighted by arrows, right) and vector tag proteins. Th-6His-tagged recombinant proteins are shown in the left panel, whereas GST-6His-tagged recombinant proteins are shown in the right panel. c Retention of PLP2 (upper panel) or RAB5 (lower panel) by a subset of deleted TPD52/Tpd52 proteins and Full-length GST-Tpd52-6His, but not vector tag proteins. Proteins detected and their molecular weights (in brackets) are shown on the right. 10 % of the SKBR-3 lysate input (SK-BR-3) was included as a loading control. Results shown are representative of those obtained in three independent experiments. (Color figure online)