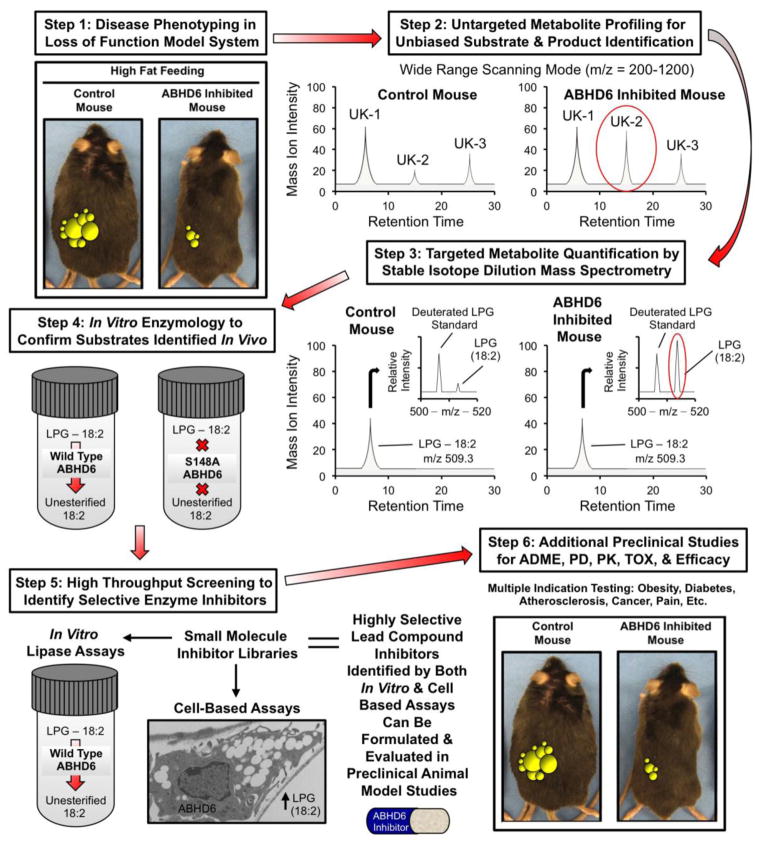
Fig. 1.
Synergistic biological and chemical methods to annotate novel lipase function. This diagram highlights the recent successful implementation of this stepwise process to annotate the previously uncharacterized lysophospholipase ABHD6. In step 1, a loss of function mouse model provides initial clues on whether enzyme inhibition would be therapeutically beneficial. For ABHD6 drug discovery, an antisense oligonucleotide (ASO) inhibition approach was taken, but knockout mouse models are the ideal screening tool. If experiments in step 1 provide support for further inquiry (ABHD6 inhibition protects against obesity, hepatic steatosis, and insulin resistance in high fat diet fed animals), steps 2 and 3 can be initiated using tissues or blood from the mice used in step 1. In step 2, lipid extracts from sources where the enzyme is abundantly expressed are unbiasedly analyzed using an untargeted LC-MS approach, where many unknown (UK-1, UK-2, and UK-3) metabolites are identified in a wide range (m/z 200–1200) scanning mode without the addition of an internal standards. The levels of unknown lipids are quantified by comparing the mass ion intensities between control vs. loss of function models. In step 3, lipid extracts from tissues or blood (where enzyme is abundantly expressed) are analyzed using a targeted quantitative approach where suspected lipid substrates are analyzed using stable isotope dilution LC-MS approaches. In both steps 2 and 3, potential enzyme substrates are expected to accumulate in loss of function mouse models, so these lipids (UK-2 or LPG-18:2) can move forward toward biochemical verification. In Step 4, the data generated in steps 2 and 3 provide clues for potential lipase substrates, and here potential substrate lipids are incubated in vitro with the purified enzyme to assay lipase activity in a reduced system. To ensure activity is intrinsic to the enzyme itself and not contaminants from the purified protein source, the same substrates are assayed with an enzyme that lacks key active site residues necessary for catalysis (S148A ABHD6). In step 5, once both in vitro and cell-based assays are developed to estimate enzyme activity; these can be used to screen small molecule inhibitor libraries to identify highly selective and biologically active lipase inhibitors. Finally, in step 6 lead small molecule inhibitors can be further evaluated for multiple indications in preclinical studies to examine ADME (absorption, distribution, metabolism, and excretion), PD (pharmacodynamics), PK (pharmacokinetics), TOX (toxicology), and efficacy.