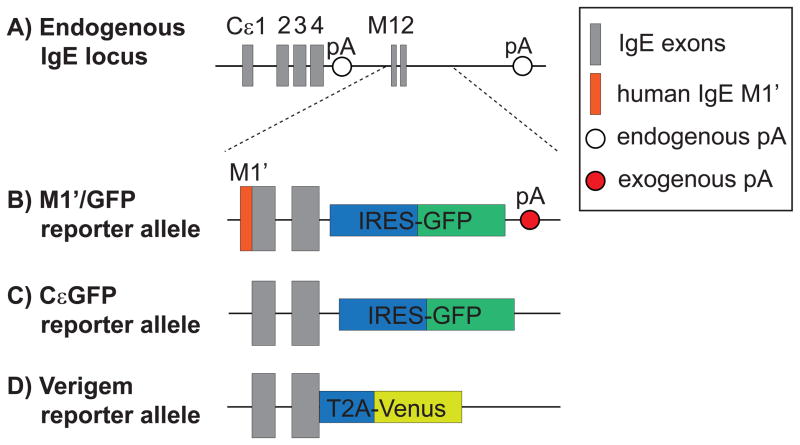
Figure 1.
Comparison of fluorescent IgE reporter alleles. (a) Structure of the native, endogenous IgE locus, which generates transcripts that encode secreted IgE versus membrane IgE (mIgE) by alternative splicing and polyadenylation. The secreted IgE transcript has a canonical polyadenylation signal sequence (pA), AAUAAA, in the intron between the CH4 and M1 exons. The mIgE transcript incorporates the M1 and M2 exons and has three potential, endogenous pAs with weak non-canonical sequences (AGUAAA, AAGAAA, or AUUAAA) [17,18]. Another possible pA with a canonical sequence is located further downstream in the intergenic region between IgE and IgA [53], although its relevance is unclear due to its long distance from the IgE M2 exon. (b and c) Schematic of the M1′/GFP and CεGFP reporter alleles, in which insertion of IRES-GFP downstream of the murine M2 exon leads to the generation a bicistronic transcript encoding mIgE and GFP. (d) Schematic of the Verigem reporter allele, in which a T2A site translationally links mIgE expression to the expression of Venus, a yellow fluorescent protein [12**]. In the M1′/GFP allele (b), the coding sequence for a 52 amino acid human M1′segment was inserted directly upstream of the murine M1 exon, and an additional exogenous pA was inserted in the 3′ UTR [11**,15]. In contrast, the CεGFP (c) and Verigem (d) reporter alleles did not introduce an exogenous pA, instead relying on the endogenous pAs in the 3′ UTR [12**,13**].