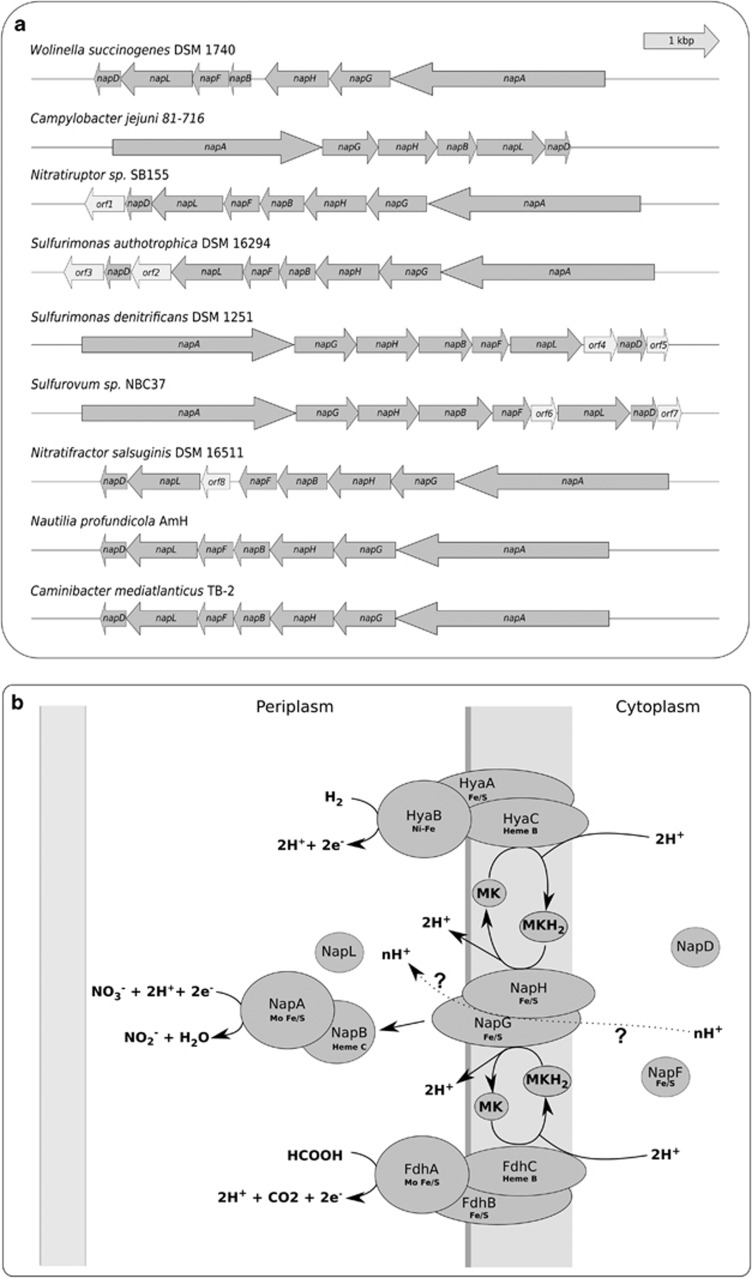
Figure 1.
(a) Organization of the nap gene cluster in different Epsilonproteobacteria. Individual genes are drawn to scale. (b) Proposed model of the Nap respiratory nitrate reduction pathway in Caminibacter mediatlanticus. Question marks denote speculative processes and the dotted line indicates the proposed contribution of the Nap complex to the generation of the proton motive force. NapAB, periplasmic nitrate reductase complex; NapHG, membrane-bound iron–sulfur complex; NapD, putative chaperone; NapL, periplasmic iron–sulfur protein; NapF, putative iron–sulfur adaptor protein; HyaA, nickel–iron-dependent hydrogenase, large subunit; HyaB, nickel–iron-dependent hydrogenanse, small subunit; HyaC, nickel–iron-dependent hydrogenase, heme-b-containing subunit; FdhA, formate dehydrogenase, molibdenum-containing large subunit; FdhB, formate dehydrogenase, small subunit; FdhC, formate dehydrogenase, heme-b-containing subunit; MK, menaquinone.