Abstract
Exploring molecular mechanisms underlying bacterial water-to-land transition represents a critical start toward a better understanding of the functioning and stability of the terrestrial ecosystems. Here, we perform comprehensive analyses based on a large variety of bacteria by integrating taxonomic, phylogenetic and metagenomic data, in the quest for a unified view that elucidates genomic, evolutionary and ecological dynamics of the marine progenitors in adapting to nonaquatic environments. We hypothesize that bacterial land colonization is dominated by a single-gene sweep, that is, the emergence of dnaE2 derived from an early duplication event of the primordial dnaE, followed by a series of niche-specific genomic adaptations, including GC content increase, intensive horizontal gene transfer and constant genome expansion. In addition, early bacterial radiation may be stimulated by an explosion of land-borne hosts (for example, plants and animals) after initial land colonization events.
Keywords: adaptive mutagenesis, bacterial land colonization, GC content, genome expansion, HGT, metagenomics
Introduction
Terrestrial ecosystems must have pressured the primordial microbial life with diverse and less movable microhabitats, scarce resources and other environmental hazards (for example, turbulence of pH and temperature, ultraviolet radiation and desiccation). Soil bacteria represent the majority of biodiversity in terrestrial ecosystems and are essentially involved in the establishment and evolution of primary elements in these ecosystems, such as carbon sequestration, nitrogen fixation and element cycling (Vogel et al., 2009; Madsen, 2011; He et al., 2012; Sanford et al., 2012; Yergeau et al., 2012). Therefore, bacterial land colonization is arguably not only one of the most challenging, but also one of the most fundamental and seminal ecological transitions in bacterial evolution. The conquest of the Earth's solid surface must have entailed significant genomic changes and genetic innovations in the overall architecture, flexible configuration and necessary elements of bacterial chromosomes to cope with this new environment. For instance, soil microbes are intensively reported to possess high metabolic versatility, such as metal-reducing ability (Venkateswaran et al., 1998), abundance of nitrous oxide reductase (Sanford et al., 2012) and antibiotic-resistant genes (Riesenfeld et al., 2004). In addition, soil-borne bacteria have been found to have the highest number of associations with diverse hosts as compared with other environment-dwelling bacteria (Hooper et al., 2009) and even the most co-occurring partners within soil environment itself (Freilich et al., 2010), together making terrestrial environment the most important yet complicated system.
The next-generation sequencing technology has made the bulk generation of pangenomic and metagenomic data sets possible for studying phylogenetic and taxonomic biogeography of soil microbial communities (Fierer and Jackson, 2006; Fierer et al., 2012a, 2012b). Currently, major efforts have been focused on investigating environment-specific genes involved in various metabolic pathways (Sanford et al., 2012; Barret et al., 2013) that are hypothesized to be responsible for bacterial niche-specific adaptations (Hacker and Carniel, 2001; Konstantinidis and Tiedje, 2005; Shapiro et al., 2009; Coleman and Chisholm, 2010). However, such investigations may only give us a glimpse of the emergence of some specific metabolic features under limited nutrients or resources instead of providing global pictures of bacterial evolution. Taking bacterial land colonization as an example, there has been limited progress in revealing how bacterial communities have conquered the land, survived in the new and harsh environments and formed unique patterns of taxonomic diversity distinct from other communities, such as the aquatic and the host associated (Madigan et al., 2011). Therefore, identification of adaptive and diversifying evolutionary processes in association with the water-to-land transition is of critical importance in systematically understanding bacterial radiation, host–pathogen coevolution and ecosystem stability.
One of our recent studies has clarified the relationship between error-prone DNA synthesis and GC content variations, showing that a paralog of replicative DnaE polymerase—DnaE2—is responsible for bacterial GC increase, whereas other mutator genes only play fine-tuning roles in this process, and that DnaE2-containing bacteria are found to be specifically enriched in soil environments (Wu et al., 2012). We conceive that DnaE2 may play a major role in the bacterial water-to-land transition, as it is more prevalent in bacterial species found in terrestrial than aquatic environments. To test this hypothesis, here we perform comprehensive comparative analyses based on a large quantity of bacteria by combining evidence from taxonomic, metagenomic and phylogenetic analyses. We also provide a unified view on bacterial land colonization by invoking GC content variation and genome size expansion.
Materials and methods
Taxonomic structure analysis
The taxonomic structure of all Proteobacteria is estimated from the National Center for Biotechnology Information (NCBI) taxonomy website (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1224). Only bacteria with available full genomic sequences are included in this analysis, that is, 1620 in total (on 19 March 2013) after the exclusion of Epsilon-proteobacteria. An alternative subset of Proteobacteria with detailed annotation of bacterial habitats and DnaE2 polymerases is collected from our previous study (195 genomes in total and 84 are DnaE2-containing bacteria) (Wu et al., 2012) and also recruited for taxonomic analysis. The data set used for estimating taxonomic structure of soil bacteria is obtained from a previous collection (Madigan et al., 2011). The presence/absence of DnaE2 in 98 randomly selected terrestrial and aquatic bacteria, which are grouped based on both taxonomic positions and lifestyles, is further reannotated (Supplementary Table S1). The detailed metadata are retrieved from Genomes Online Database (http://www.genomesonline.org/; Pagani et al., 2012).
Detection of DnaE2 in metagenomic data sets
The metagenomic data for the Sargasso Sea are collected from a previous publication (Venter et al., 2004). The newly released metagenomic data from Peru Margin sediment are also collected (Orsi et al., 2013). All other metagenomic data are from JGI metagenomics program (http://genome.jgi.doe.gov/programs/metagenomes/index.jsf). We collect 7 334 298 protein sequences from aquatic environment (including marine and fresh water) and 14 303 396 protein sequences from soil environment for the detection of DnaE polymerases. The detailed identification procedures are described in (Supplementary Figure S1). We first extract all peptide sequences annotated as ‘DNA polymerase III alpha subunit' (DnaE/PolC polymerases) with length of >100 amino acids and then recruit these candidate DnaE/PolC sequences for further HMM (Hidden Markov Model) profiling using HMMER (version 3.0) (Finn et al., 2011). Three different DnaE/PolC HMM profiling matrices (from DnaE1+DnaE3, DnaE2 and PolC, respectively) are built (hmmbuild) based on our previous curation (Wu et al., 2012). These three HMM matrices are then used to search each candidate DnaE polymerase extracted from the metagenomic data sets (hmmsearch) ending with three different HMM profiling scores and relative E-values. The scores reflecting the identity of the candidate sequence with each of the three HMM profiling matrices are then compared for further classification. The candidate sequence is identified as DnaE2 only when its identity with DnaE2 HMM matrix is the largest and meantime 1.5 times larger than that with the DnaE1+DnaE3 HMM matrix. The DnaE1+DnaE3 polymerase is also identified in the same way (as long as the score with DnaE1+DnaE3 is the largest and also 1.5 times larger than the score with the DnaE2 HMM matrix), whereas a polymerase is identified as PolC as long as it has the best similarity score with PolC HMM matrix, as PolC is very distinct from other polymerases. All other polymerases are grouped as unclassified for better data quality and excluded from further analysis. Three mutator genes (mutT, mutY and mutM) that participate in DNA repair and contribute to GC content variation (loss of mutT increases GC, whereas loss of mutY/M increases AT) (Garcia-Gonzalez et al., 2012; Wu et al., 2012) are also collected in order to examine their relative abundances in terrestrial versus aquatic environment. As these repair genes tend to have shorter lengths (encoding ∼200 amino acids), only sequences with HMM scores ⩾50 and E-value ⩾1e−10 are extracted for further analysis. Besides, DnaE sequences in 12 freshwater samples and 20 FACE (Free-Air Carbon Dioxide Enrichment) soil samples are used for sequencing saturation analysis and the relative abundance of DnaE2 in six samples of different depths (5, 30, 50, 70, 91, and 159 m) from Peru Margin marine sediment are also examined. The proportion of DnaE2 (DnaE2%) is calculated as follows (based on the fact that DnaE1+DnaE3 sequences are generally single copied and thus their numbers can roughly reflect the total number of bacteria):
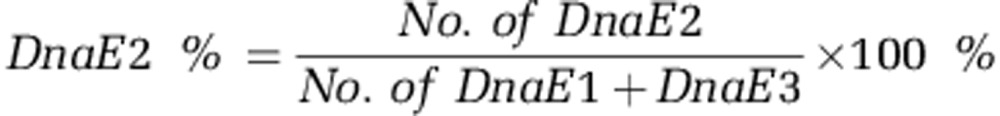 |
Phylogenetic tree construction
We build all phylogenetic trees using MEGA 5.05 (Tamura et al., 2011) under JTT+Γ4 model. The phylogeny of 137 DnaE2 sequences is built using neighbor-joining method with 100 bootstraps. A total of 10 random selected DnaE1 sequences from Actinobacteria is used as the outgroup. The main topology of DnaE2 tree is also validated by a more comprehensive data set of outgroup including 25 DnaE1 sequences from five different taxonomic groups using both neighbor joining (Supplementary Figure S2A) and more robust maximum likelihood methods (Supplementary Figure S2B). Four DnaE2 sequences (one from Nitrospirae, one from Verrucomicrobia and two from Delta Proteobacteria) identified previously by HMM profiling method are excluded for further analysis as they are phylogenetically classified as DnaE1 (Supplementary Figure S2). The phylogenies of three case studies referring to Chlamydiae-Verrucomicrobia, Azospirillum (Alpha Proteobacteria) and Beta Proteobacteria are built by DnaE1 sequences using neighbor-joining method with 500 bootstraps and visualized with the help of the online iTOL tool (Letunic and Bork, 2011).
Results
Evidence from the taxonomic structure
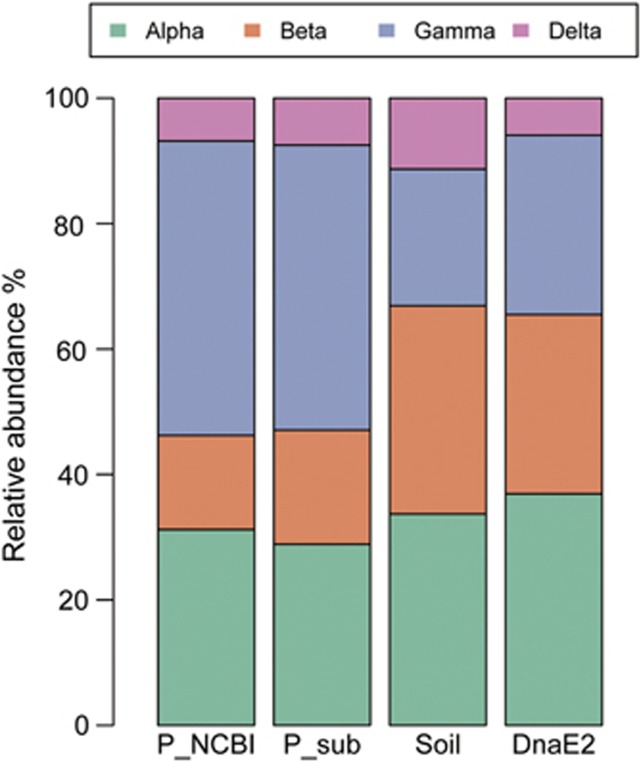
Diverse soil bacterial communities are often characterized by the dominance of Proteobacteria (mainly within Gamma, Beta and Alpha class), Actinobacteria, Acidobacteria, Planctomycetes and Verrucomicrobia (Dedysh et al., 2006; Zhou et al., 2009; Bergmann et al., 2011; Montana et al., 2012). Community-based analyses in paleosols also confirm this distinct taxonomic distribution (Chandler et al., 1998; Hart et al., 2011). Coincidently, we notice that these dominant bacterial phyla in soil are, intriguingly, also DnaE2 bearing, with very few exceptions (Table 1). For further validation, we compare the taxonomic structure of DnaE2-containg Proteobacteria with that of terrestrial Proteobacteria (given that Proteobacteria contain the most available genome sequences representing an unbiased sampling). Our result demonstrates that these two groups of bacteria have very similar taxonomic structure—both underrepresentation of Gamma-proteobacteria but overrepresentation of Beta-proteobacteria, whereas the proportions of Alpha- and Delta-proteobacteria are not much variable as compared with the entire Proteobacteria population (Figure 1). Interestingly, Epsilon-proteobacteria are undetected or nearly absent in either data sets. The similar community structure between DnaE2-bearing and soil-dwelling bacteria strongly indicates that the appearance of dnaE2 plays an important role in shaping the biogeographic pattern of terrestrial bacteria. Moreover, the presence of DnaE2 in most bacteria is associated with terrestrial environment, whereas the absence of DnaE2 is found to be common in aquatic bacteria even when comparing within the same phylum (Supplementary Table S1), suggesting that it is DnaE2 not taxonomy that is linked to environmental adaptations.
Table 1. Number of DnaE2-containing bacteria in each phylum.
| Group | Bacteria | Number of dnaE2 bacteria |
|---|---|---|
| Terrabacteria | Actinobacteria | 202 |
| Chlorofexi | 4 | |
| Cyanobacteria | 0 | |
| Deinococcus | 3 | |
| Firmicutes | 5 | |
| Hydrobacteria | Acidobacteria | 6 |
| Aquificae | 0 | |
| Bacteroidetes | 9 | |
| Chlamydiae | 0 | |
| Chlorobi | 0 | |
| Fusobacteria | 0 | |
| Gemmatimonadetes | 1 | |
| Planctomycetes | 6 | |
| Proteobacteria | 369 | |
| Spirochaetes | 0 | |
| Synergistetes | 0 | |
| Tenericutes | 0 | |
| Thermotogae | 0 | |
| Verrucomicrobia | 5 |
Bacteria in bold are the DnaE2-containing bacteria.
Figure 1.
Taxonomic structure of DnaE2-containing and soil-dwelling bacteria. ‘P_sub' stands for a subset of Proteobacteria collected from our previous study (Wu et al., 2012) that in general reflects the taxonomy structure of all sequenced Proteobacteria in the NCBI database (P_NCBI). The data set for the ‘Soil' bacteria is from Madigan et al. (2011). ‘DnaE2' includes all DnaE2-containing bacteria in ‘P_sub'.
Evidence from metagenomic study
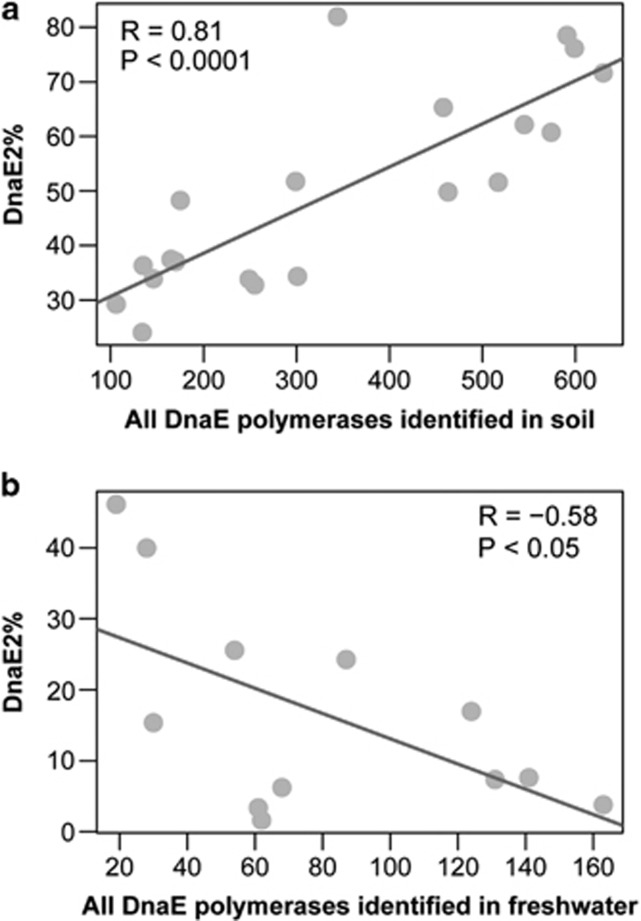
We further explore the abundance of DnaE2 using the abundant metagenomic data, as metagenomes enable systematic understanding of gene content, functional relevance and genomic plasticity in natural microbial communities. We argue that DnaE2 is more prevalent among terrestrial than aquatic bacteria and the presence of DnaE2 contributes considerably to the success of bacterial land colonization. To test this proposition, we collect six different metagenomic samples: three data sets generated from aquatic environment DNA and three from terrestrial DNA. Consistent with our expectations, a clear enrichment of DnaE2 was identified in terrestrial (∼55–68% DnaE2-containing bacteria) than in aquatic (only ∼11–21% DnaE2-containing bacteria) environments (Table 2). We also detect the proportion of DnaE2-containing bacteria in each of the 12 freshwater and 20 FACE soil samples. Intriguingly, we find that the sequencing depth (indicated by the abundance of DnaE polymerases) correlates linearly with the proportion of DnaE2 (Figure 2). Specifically, soil samples present a strong upward trend, implying that the proportion of DnaE2-containing bacteria may constitute >70% of total bacterial species at least in the FACE soil samples (R=0.81, P<0.0001), whereas freshwater samples exhibit a strong descending trend, indicating that DnaE2-containing bacteria in this environment are clearly underrepresented (as low as ∼10% R=−0.58, P<0.05). Taken together, these results clearly suggest that dnaE2 is one key soil-specific gene. In addition, we notice that the lower GC contents (∼48%) of bacteria found in the upper level of the marine sediment (at depths of 5 and 30 m) are associated with lower abundance of DnaE2 (on average, only 42.4% are DnaE2-containing bacteria), as compared with the deep marine sediment below 50 m (at depths of 50, 70, 91 and 159 m) where higher GC contents (∼53.5%) are found to be correlated with higher proportion of DnaE2-containing bacteria (84.5%) (Supplementary Table S2), confirming the major role of DnaE2 to bacterial GC increase. We can only roughly estimate the relative abundance of three mutator genes because of lack of benchmarking gene of similar length (Supplementary Table S3). The results indicate that in all three samples of aquatic environment, mutT is indeed more abundant than mutY/M. But in terrestrial environment, only sample from FACE site has a slightly higher enrichment of mutY/M than mutT, whereas the other two samples unexpectedly have more mutT genes. Currently, we are not sure whether this unexpected pattern in two soil samples is a result of lower level of sequencing depths or alternatively it stands for a common observation for genes that are playing subsidiary roles in altering genomic GC contents.
Table 2. Summary of DnaE and PolC polymerases (⩾100 amino acids) in six metagenomic samples.
| Samples | DnaE1+DnaE3 | DnaE2 | PolC | unclassified | Total | DnaE2% |
|---|---|---|---|---|---|---|
| North Pacific Ocean | 1414 | 307 | 209 | 543 | 2473 | 21.71 |
| Sargasso sea | 1353 | 283 | 798 | 110 | 2544 | 20.92 |
| Fresh watera | 872 | 96 | 85 | 388 | 1441 | 11.01 |
| Peru Margin sedimentb | 92 | 52 | 29 | 19 | 192 | 56.52 |
| Minnesota Farm | 66 | 45 | 5 | 40 | 156 | 68.18 |
| FACEc | 4406 | 2450 | 393 | 2034 | 9283 | 55.61 |
The number of polymerases is listed in each group for each sample.
Twelve samples from fresh water are all recruited for further data saturation analysis in Figure 2.
Six samples of marine sediment of different depths are further compared in order to clarify the association between DnaE2% and GC content variations in Supplementary Table S2.
Twenty samples from FACE (Free-Air Carbon Dioxide Enrichment) sites are also recruited for further data saturation analysis in Figure 2.
Figure 2.
Correlation between DnaE2 (%) and sequencing depth. The total number of DnaE sequences (DnaE1+DnaE2+DnaE3) and the proportion of DnaE2-containing bacteria (DnaE2%) are indicated in the x and y axes, respectively, and the former can be roughly used as a measure of sequencing depths. There are 20 soil (a) and 12 freshwater (b) samples recruited for this analysis.
Evidence from phylogenetic analysis
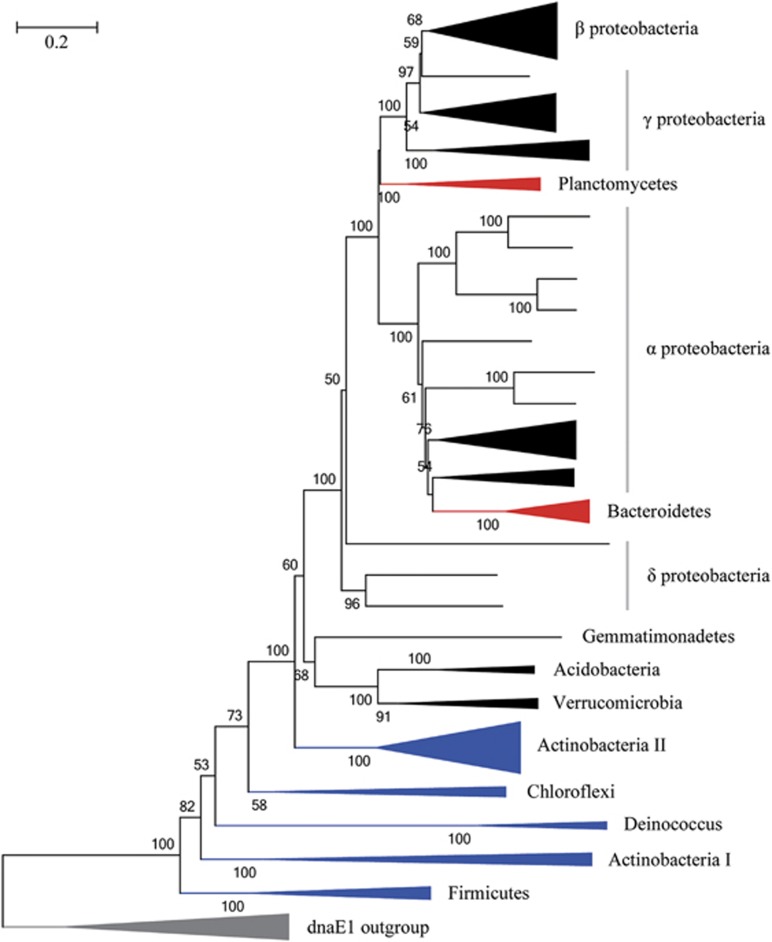
We also construct the phylogenetic tree of DnaE2 polymerase (Figure 3) using 10 DnaE1 sequences as an outgroup. We find that dnaE2 is often involved in horizontal gene transfer, for example, dnaE2 of bacteria in Planctomycetes and Bacteroidetes. However, the most striking finding is that DnaE2 in terrabacteria (that is, Actinobacteria, Firmicutes, Chloroflexi and Deinococcus-Thermus) (Battistuzzi et al., 2004; Battistuzzi and Hedges, 2009) are more closely related to DnaE1, implying that dnaE2 might first appear in terrabacteria, which further confirms our idea about the unrecognized outstanding contribution of DnaE2 to bacterial water-to-land transition.
Figure 3.
Phylogenetic tree of DnaE2. The tree is constructed by using MEGA 5.0 under JTT+Γ4 model (with 100 bootstraps). Ten randomly selected DnaE1 sequences from Actinobacteria are used as outgroup (in gray color). Terrabacteria are colored in blue. Horizontally transferred DnaE2 sequences are colored in red. The main topology of DnaE2 tree is validated by a more comprehensive data set of outgroup using both neighbor-joining (NJ) and maximum likelihood (ML) methods (Supplementary Figure S2).
Case studies
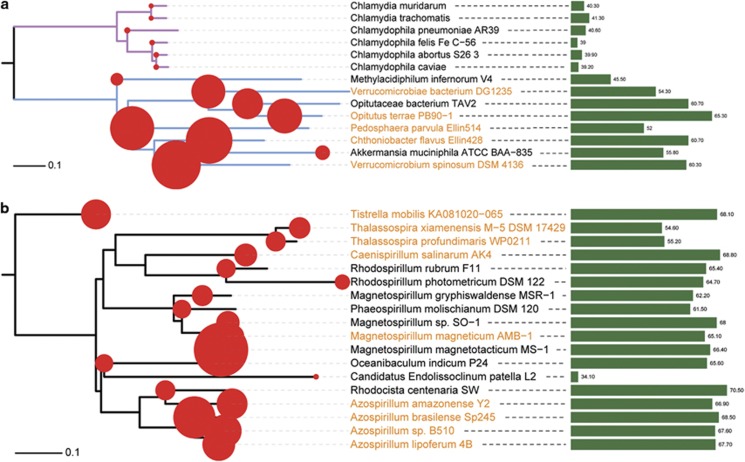
Chlamydiae and Verrucomicrobia are known to be closely related (Wagner and Horn, 2006; Griffiths and Gupta, 2007), but it remains unclear why they have distinct ecological niches and genomic features (for example, GC contents, genome sizes). We thus use them for a further dissection of the enigmatic relationship among GC content variation, genome size expansion and ecological shifts. Our comparative analysis reveals that most of the Verrucomicrobia bacteria have gained dnaE2 gene, presumably provoking higher GC content and better fitness in soil environment. In contrast, Chlamydiae, lacking dnaE2, are evolving toward a very different destiny, namely, they are involved in a series of ecological shifts (for example, from environment to animals or from animals to humans) and host–pathogen coevolution events (Horn et al., 2004; Roulis et al., 2012), followed by dramatic genome reduction and GC decrease (Figure 4a). Notably, there are only three bacteria (Methylacidiphilum infernorum V4, Akkermansia muciniphila and Opitutaceae bacterium TAV2) in Verrucomicrobia found to have lost dnaE2. The absence of dnaE2 in the first two bacteria can be better indicated by their dramatic genome reductions, and the loss of dnaE2 in Opitutaceae bacterium TAV2 (or Diplosphaera colitermitum TAV2) still needs further examination owing to its incomplete genome sequence.
Figure 4.
The gain and loss of dnaE2 and its correlation with bacterial land colonization. Bacteria in Chlamydiae-Verrucomicrobia (a, with branches colored in magenta and light blue) and Rhodospirillaceae (b) are used for this case study. The phylogenetic trees are constructed by using MEGA 5 under JTT+Γ4 model (with 500 bootstraps). Red circles mapped on the trees are proportional to the genome size of each bacterium. Bacteria names labeled in brown color stand for DnaE2-containing bacteria and the green bars in the right panel are proportional to the GC content of each bacterium. Although the non-Azospirillum bacteria are correlated with aquatic environment, most of them actually dwell in the boundaries or mixtures of soil and water, such as water sediment (for example, M. magnetotacticum, M. gryphiswaldense and T. profundimaris) and ditch mud (for example, R. rubrum and P. molischianum).
We also examine bacteria of the genus Azospirillum that are reported to have transitioned from marine to terrestrial environments (Wisniewski-Dyé et al., 2011). Our results demonstrate that all the four soil-dwelling bacteria of this genus, having high GC contents and large genome sizes, are indeed DnaE2-containing bacteria (Figure 4b). However, we find it is very unconvincing that bacterial land colonization may begin from this genus (Wisniewski-Dyé et al., 2011), given the fact that Alpha-proteobacteria are abundant in soil. As in this case, we notice that most bacteria of the non-Azospirillum within the Rhodospirillaceae family have comparable high GC contents with that of the four bacteria in Azospirillum (except Candidatus Endolissoclinum patella L2 that has experienced striking genome reduction because of its ancient symbiotic relationship with marine tunicate; Kwan et al., 2012). Therefore, we infer that dnaE2 should not only appear in Azospirillum but also be common among non-Azospirillum bacteria. Our genome screening indeed show that at least five of these non-Azospirillum bacteria also possess dnaE2, and thus we further wonder why these water-associated non-Azospirillum bacteria (Wisniewski-Dyé et al., 2011) are DnaE2-containing bacteria if DnaE2 is the decisive element of bacterial land colonization. Our following ecological survey in the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) microbial collections (http://www.dsmz.de/catalogues/catalogue-microorganisms.html) indicates that although most of the non-Azospirillum bacteria are associated with aquatic environments, they actually dwell in the boundary zones between water and soil environments, such as water sediment (for example, Magnetospirillum magnetotacticum, M. gryphiswaldense and Thalassospira profundimaris) and ditch mud (for example, Rhodospirillum rubrum and Phaeospirillum molischianum). In addition, Tistrella bauzanensis that forms a coherent cluster with DnaE2-bearing T. mobilis is reported to be soil dwelling (Zhang et al., 2011). Therefore, we argue that not only the Azospirillum genus, but also bacteria of the entire Rhodospirillaceae family should have already begun their journey toward land colonization.
Discussion
The gain and loss of dnaE2 and its association with bacterial land colonization
We have reported previously that ∼68% of the terrestrial bacteria are DnaE2-containing bacteria (Wu et al., 2012). Here we combine evidence from taxonomic, metagenomic and phylogenetic analyses, revealing that the emergence of dnaE2 is a key genetic innovation underlying the success of bacterial land colonization. Given the strong correlation between dnaE2-bearing and soil-dwelling bacteria, we propose that bacterial land colonization is an ongoing process occurring successfully only when the bacterium acquires a dnaE2 gene and has nothing to do with taxonomic affiliations. This inference explains why the water-to-land transition of the Azospirillum genus (Wisniewski-Dyé et al., 2011) has occurred much later than the suggested divergence of hydrobacteria and terrabacteria (Battistuzzi et al., 2004; Battistuzzi and Hedges, 2009). The horizontal transfer of dnaE2 to Azospirillum genus should have occurred much later, potentially consistent with the radiation of vascular plants on land as suggested by Wisniewski-Dyé et al. (2011), whereas terrabacteria might be one of the first groups of bacteria that has gained dnaE2. Our results also provide insights into the strikingly different evolutionary scenarios of the two closely related groups—Chlamydiae and Verrucomicrobia. The ancestor of Verrucomicrobia has gained dnaE2 followed by GC increase, genome expansion and land colonization, whereas the dnaE2-deficient early lineages of Chlamydiae have built an ancestral relationship with diverse hosts and experienced dramatic genomic reduction. Sphaerobacter thermophilus in Chloroflexi is also found to be very different from its close relatives by having higher GC content and living in terrestrial environment (originally isolated from sewage sludge; Pati et al., 2010). Our result from genome analysis indicates that this bacterium also possess dnaE2. The only exception is Cyanobacteria that belong to terrabacteria yet with no evidence of possessing dnaE2. However, we believe one of the earliest lineage of Cyanobacteria may once have had dnaE2 and conquered the land environment as evidenced by current phylogenetic analyses that tend to root Cyanobacteria at Gloeobacter violaceus (Nakamura et al., 2003), a high GC bacterium that prefers terrestrial environment (SÁNchez-Baracaldo et al., 2005). That is to say, the current marine habitant of Cyanobacteria may actually be a back-to-the-sea event, which is in good agreement with previous studies (SÁNchez-Baracaldo et al., 2005; Wisniewski-Dyé et al., 2011).
GC increase vs genome expansion in relation to environmental adaptations
We have provided evidence in our previous (Wu et al., 2012) and present studies to support the proposition that DnaE2 plays the major role in a ‘dice-casting' of bacterial evolution, although debatable in detailed molecular mechanisms of bacterial GC increase. We further argue that GC increase is the prerequisite of genome expansion based on the following grounds. First, it has long been known that GC content increase is linearly correlated with genome size expansion (Musto et al., 2006). Second, GC content is well recognized as one of the important barriers for horizontal gene transfers in recent studies as bacterial genic GC contents have not much deviated from the GC content of the main chromosomes (Popa et al., 2011; Nishida, 2012a, 2012b; Hayek, 2013). Third, bacteria can selectively silence foreign genes whose GC content is lower than the host genomic GC content (Navarre et al., 2006; Navarre et al., 2007). Fourth, according to the phylogenetic analysis of dnaE2, this gene may first appear in terrabacteria that are estimated to have begun colonizing land as early as 3.54 to 2.83 Gyr (Battistuzzi and Hedges, 2009). Therefore, there is a possibility that the emergence of dnaE2 (and subsequent GC increase) has happened before the burst of de novo gene-family birth (∼3.33–2.85 Gyr) or at least at the early stage of this ‘Archaean genetic expansion' when extensive genome expansions have not been summoned (David and Alm, 2011). Taken together, the gain of exogenous environmental DNA/genes of high GC content rarely happens without an initial GC content increase of the host genome.
Genome expansion stimulated by GC increase enables the success of bacterial land colonization by providing bacteria with higher rate of distant horizontal transfers from donors of similar genome sizes (Cordero and Hogeweg, 2009) and with more genes involved in regulation, signaling or secondary metabolism (Cases et al., 2003; Konstantinidis and Tiedje, 2004). Within this context, we can better understand why environment pressures are misleadingly reported to shape bacterial GC contents based on the finding that terrestrial bacteria generally have higher GC content than aquatic bacteria (Foerstner et al., 2005). In addition, we note that the RcGTA (Rhodobacter capsulatus gene transfer agent) (Lang and Beatty, 2007) that has been reported to be able to boost the efficiency of horizontal gene transfers (McDaniel et al., 2010) is enriched in the soil than in the marine environments (Supplementary Figure S3), providing additional evidence to explain bacterial genome expansion in terrestrial environment.
Based on the immense metabolic flexibility of high-GC DnaE2-containing bacteria, we conclude that there must be ample unusual metabolic features because of their enhanced ability to gain exogenous genetic materials. To take Symbiobacterium thermophilum as an example, this DnaE2-containing bacterium is known to possess a variety of respiratory systems found only in Gram-negative bacteria (Ueda et al., 2004); DnaE2-containing Silicibacter pomeroyi is also found to have some unusual metabolic pathways to deal with nutrient-poor habitats (Moran et al., 2004); there is even one DnaE2-containing bacterium reported to have some eukaryotic features, for example, genes involved in sterol synthesis (Pearson et al., 2003). In addition, most bacteria armed with compound-degrading ability are also revealed to be DnaE2-containing bacteria (Phale et al., 2007; Wu et al., 2012), uncovering their great roles in bioremediation. Furthermore, not only unusual but also novel metabolic pathways tend to be found in DnaE2-containing bacteria (Table 3). Thus, it is not surprising to detect a new pathway for calcification in Cyanobacteria (Couradeau et al., 2012) as postulated by our inference that Cyanobacteria may once have possessed dnaE2 and dwelt in terrestrial environment. Even genes involved in oxygenic photosynthesis that are often involved in horizontal gene transfer (Shi and Falkowski, 2008) may also originate during terrestrial adaptation (Battistuzzi et al., 2004).
Table 3. Examples of new metabolic pathways identified in the DnaE2-containing bacteria.
| DnaE2 bacteria | Phylum | New pathways | Reference |
|---|---|---|---|
| Burkholderia cenocepacia | Actinobacteria | Anoxic persistence | Sass et al. (2013) |
| Methylococcus capsulatus | α-Proteobacteria | Gluconeogenesis; the ability to use copper in regulation of methanotrophy; sterol and hopanoid biosynthesis | Ward et al. (2004) |
| Ruegeria pomeroyi | α-proteobacteria | Assimilation of dimethylsulfoniopropionate | Reisch et al. (2011) |
| Thermomicrobium roseum | Chloroflexi | Oxidization of CO aerobically | Wu et al. (2009) |
| Candidatus Methylomirabilis oxyfera | Division NC10 | Methane oxidation under anoxic conditions | Ettwig et al. (2010) |
| Pseudomonas sp. strain MT1 | γ-Proteobacteria | 4- and 5-Chlorosalicylate degradation | Nikodem et al. (2003) |
| Gemmatimonas aurantiaca | Gemmatimonadetes | Synthesis of the carotenoids | Takaichi et al. (2010) |
A unified view of bacterial land colonization
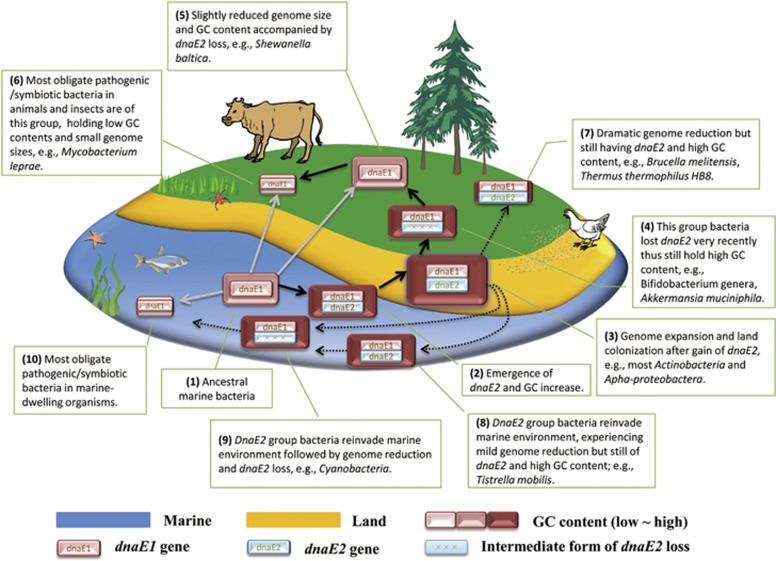
Our hypothesis, for a better display, is illustrated in Figure 5. The marine ancestral dnaE1-containing bacteria (step 1) gain an active copy of dnaE2 (evolved from a dnaE1 duplicate) followed by GC increase (step 2), genome expansion and land colonization (step 3). Starting from step 3, there are three different possible evolutionary scenarios. First, some bacteria in this stage further experience niche-restricted adaptations because of ecological shifts (to mammals for example), lose dnaE2 (step 4) and continue to evolve generally in the form of GC decrease (step 5) and genome reduction (step 6) owing to host jump (for example, from environment to insects). Second, there may be also some that have experienced striking genome reduction but still possess dnaE2 and thus keep high GC content (step 7). Third, others possibly reinvade the marine environment (step 8 and 9) and further spread to marine-borne hosts (step 10). In addition, pathogenic/symbiotic bacteria in various land- and water-dwelling organisms may also evolve from lineages derived directly from ancestral dnaE1-containing bacteria.
Figure 5.
A unified model of bacterial land colonization. We propose a conceptual framework here to help clarify the emergence of dnaE2 and its contribution to bacterial land colonization through a series of genomic changes including GC content increase and genome expansion. Marine-borne (for example, seaweed and fish) and land-borne organisms (for example, trees for plants, beetle for insects and cattle for mammals) are also illustrated here for exemplification of bacterial radiation because of host diversification. The dnaE-containing rectangles stand for different stages of bacterial evolution with the rectangle sizes proportional to bacterial genome sizes. There are mainly three routes for bacterial evolution. The first route is bacterial shifts from ocean to land owing to emergence of dnaE2, GC increase and genome expansion (black arrows) and further radiations and adaptations because of the explosion of diverse land hosts. The second route is the reinvasion of the marine bacteria after land colonization (black dashed arrows). The last route for bacterial host jumps is directly from oceanic to land-borne organisms (gray arrows). Key genomic innovations and bacterial examples are labeled for each stage (up to 10).
In summary, the bacterial water-to-land transition is characterized by a new pathway of adaptive mutagenesis that arms the bacteria with a genome of higher GC content, larger genome size and an open pan-genome so that they have better ability to deal with strange and hostile soil environments. This new pathway of adaptive mutagenesis or genetic innovation recruits the coding product of dnaE2 that may come from dnaE1 gene duplication for error-prone DNA synthesis (observed as GC increase). The genomic GC increase shaped by DnaE2-invloved error-prone DNA repair is then inferred to be the prime for subsequent bacterial genome expansion that in return confers bacteria with a fitness advantage in the soil-based environment. Driven by positive selection, dnaE2 passes through one bacterium to another (by horizontal gene transfer or recombination), sweeping through different bacterial phyla and triggering new ecological differentiations. This view is consistent with the model of ‘ecotype-formation mutations' (Cohan and Perry, 2007). However, significant work remains to be done in order to reveal the detailed genetic basis of these adaptive evolutionary changes.
Further radiation of the soil microbial community
Based on this conceptual framework, we also find clues on the radiation of soil-dwelling microbial communities to other land hosts. For instance, the smallest Beta-proteobacterial bacterium Candidatus Glomeribacter gigasporarum (∼1815 genes), which has a surprisingly high GC content (54.8%) (Ghignone et al., 2012), is found to maintain an ancient relationship with arbuscular mycorrhizal fungi (Jargeat et al., 2004). We infer that this bacterium may once have possessed dnaE2 and experienced ecological shift from soil to fungi and dramatic genome reduction because most of its relatives are dnaE2-containing and soil-dwelling bacteria (Supplementary Figure S4). This argument is also supported by another closely related fungal endosymbiont, that is, Burkholderia rhizoxinica HKI 454, that is still holding dnaE2 and thus higher GC content (60.7%) because of a more modest genome reduction (∼3936 genes) compared with Candidatus Glomeribacter gigasporarum. In addition, recent metagenomic studies (Cazemier et al., 1999; Cazemier et al., 2003; Ventura et al., 2007; Kaltenpoth, 2009; Salem et al., 2012; Sudakaran et al., 2012) have repeatedly revealed that some Actinobacteria play essential roles in insect gut by helping their insect hosts to use diverse plant-fiber-derived polysaccharides. As most Actinobacteria are DnaE2-containing and particularly widespread in the terrestrial environment, they are supposed to transit from soil to plant host and therefore are regularly encountered by soil-dwelling insects feeding on plants (Kaltenpoth, 2009). There may also be some Actinobacteria that have experienced soil-to-human host jump, for example, Turicella otitidis, a DnaE2-containing human pathogen in skin and ear that has a small genome (∼1800 genes) but high GC content (∼71%) (Brinkrolf et al., 2012). In addition, the shared antibiotic resistome of soil bacteria and human pathogens (Forsberg et al., 2012) can also support the pivotal roles of soil bacterial communities to the bacterial radiation. Furthermore, we infer that it is an easier shift for bacteria to move from soil to inland fresh water than from soil back to marine as evidenced by limited overlaps between freshwater and marine microbial communities (for example, soil abundant Beta-proteobacteria are also typical freshwater goers yet nearly completely absent in the oceans) (Philippot et al., 2010).
Conclusion
Here we perform comprehensive analyses and try to put together a unified view on bacterial land colonization that has been proposed to involve gene duplication, function diversification and a series of genomic alternations that include GC content increase and genome expansion. We report that the emergence and the sweep of dnaE2 are the key genomic innovations underlying bacterial adaptation to terrestrial environment based on three lines of evidence. First, the similar taxonomic structure between dnaE2-containing and soil-dwelling bacteria implies that dnaE2 plays a decisive role in shaping the unique biogeographic pattern of soil microbial community. Second, metagenomic data screening reveal that dnaE2 is indeed soil specific. Third, phylogenetic analyses indicate that dnaE2 may first appear in terrabacteria. Taken together, these results consistently and clearly show that dnaE2 is of great relevance to the success of terrabacterial land colonization, providing a new perspective for the study of bacterial radiation after land colonization. Future studies will be focused on experimental validation of this genome-based hypothesis.
Acknowledgments
We thank Dr Nicolás Pinel for providing the data set for analyzing the taxonomic structure of the terrestrial bacteria and Dr Nicolas Galtier and Dr Konstantinos T Konstantinidis for helpful and constructive discussions on this work. We also thank three anonymous reviewers for providing helpful comments on our manuscript. This work was supported by grants from the ‘100-Talent Program' of Chinese Academy of Sciences (Y1SLXb1365 to ZZ), National Programs for High Technology Research and Development (863 Program; 2012AA020409 to ZZ) and the Special Foundation Work Program (2009FY120100 to JY), the Ministry of Science and Technology of the People's Republic of China.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Barret M, Egan F, O'Gara F. Distribution and diversity of bacterial secretion systems across metagenomic datasets. Environ Microbiol Rep. 2013;5:117–126. doi: 10.1111/j.1758-2229.2012.00394.x. [DOI] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Hedges SB. A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol. 2009;26:335–343. doi: 10.1093/molbev/msn247. [DOI] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkrolf K, Schneider J, Knecht M, Ruckert C, Tauch A. Draft genome sequence of Turicella otitidis ATCC 51513, isolated from middle ear fluid from a child with otitis media. J Bacteriol. 2012;194:5968–5969. doi: 10.1128/JB.01412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I, de Lorenzo V, Ouzounis CA. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003;11:248–253. doi: 10.1016/s0966-842x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- Cazemier AE, Verdoes JC, van Ooyen AJ, Op den Camp HJ. Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae. Appl Environ Microbiol. 1999;65:4099–4107. doi: 10.1128/aem.65.9.4099-4107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazemier AE, Verdoes JC, Reubsaet FA, Hackstein JH, van der Drift C, Op den Camp HJ. Promicromonospora pachnodae sp. nov., a member of the (hemi)cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Antonie Van Leeuwenhoek. 2003;83:135–148. doi: 10.1023/a:1023325817663. [DOI] [PubMed] [Google Scholar]
- Chandler DP, Brockman FJ, Bailey TJ, Fredrickson JK. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb Ecol. 1998;36:37–50. doi: 10.1007/s002489900091. [DOI] [PubMed] [Google Scholar]
- Cohan FM, Perry EB. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. The impact of long-distance horizontal gene transfer on prokaryotic genome size. Proc Natl Acad Sci USA. 2009;106:21748–21753. doi: 10.1073/pnas.0907584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couradeau E, Benzerara K, Gerard E, Moreira D, Bernard S, Brown GE, Jr., et al. An early-branching microbialite cyanobacterium forms intracellular carbonates. Science. 2012;336:459–462. doi: 10.1126/science.1216171. [DOI] [PubMed] [Google Scholar]
- David LA, Alm EJ. Rapid evolutionary innovation during an Archaean genetic expansion. Nature. 2011;469:93–96. doi: 10.1038/nature09649. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol. 2006;72:2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA. 2012;109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerstner KU, von Mering C, Hooper SD, Bork P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005;6:1208–1213. doi: 10.1038/sj.embor.7400538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38:3857–3868. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez A, Rivera-Rivera RJ, Massey SE. The presence of the DNA repair genes mutM, mutY, mutL, and mutS is related to proteome size in bacterial genomes. Front Genet. 2012;3:3. doi: 10.3389/fgene.2012.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L, et al. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J. 2012;6:136–145. doi: 10.1038/ismej.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. Phylogeny and shared conserved inserts in proteins provide evidence that Verrucomicrobia are the closest known free-living relatives of chlamydiae. Microbiology. 2007;153:2648–2654. doi: 10.1099/mic.0.2007/009118-0. [DOI] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart KM, Szpak MT, Mahaney WC, Dohm JM, Jordan SF, Frazer AR, et al. A bacterial enrichment study and overview of the extractable lipids from paleosols in the Dry Valleys, Antarctica: implications for future Mars reconnaissance. Astrobiology. 2011;11:303–321. doi: 10.1089/ast.2010.0583. [DOI] [PubMed] [Google Scholar]
- Hayek N. Lateral transfer and GC content of bacterial resistant genes. Front Microbiol. 2013;4:41. doi: 10.3389/fmicb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Piceno Y, Deng Y, Xu M, Lu Z, Desantis T, et al. The phylogenetic composition and structure of soil microbial communities shifts in response to elevated carbon dioxide. ISME J. 2012;6:259–272. doi: 10.1038/ismej.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SD, Mavromatis K, Kyrpides NC. Microbial co-habitation and lateral gene transfer: what transposases can tell us. Genome Biol. 2009;10:R45. doi: 10.1186/gb-2009-10-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, et al. Illuminating the evolutionary history of chlamydiae. Science. 2004;304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- Jargeat P, Cosseau C, Ola'h B, Jauneau A, Bonfante P, Batut J, et al. Isolation, free-living capacities, and genome structure of “Candidatus Glomeribacter gigasporarum,” the endocellular bacterium of the mycorrhizal fungus Gigaspora margarita. J Bacteriol. 2004;186:6876–6884. doi: 10.1128/JB.186.20.6876-6884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenpoth M. Actinobacteria as mutualists: general healthcare for insects. Trends Microbiol. 2009;17:529–535. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci USA. 2012;109:20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Stahl DA, Clark DP.2011Brock Biology of Microorganisms13th edn.Benjamin Cummings: San Francisco [Google Scholar]
- Madsen EL. Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol. 2011;22:456–464. doi: 10.1016/j.copbio.2011.01.008. [DOI] [PubMed] [Google Scholar]
- McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. High frequency of horizontal gene transfer in the oceans. Science. 2010;330:50. doi: 10.1126/science.1192243. [DOI] [PubMed] [Google Scholar]
- Montana JS, Jimenez DJ, Hernandez M, Angel T, Baena S. Taxonomic and functional assignment of cloned sequences from high Andean forest soil metagenome. Antonie Van Leeuwenhoek. 2012;101:205–215. doi: 10.1007/s10482-011-9624-8. [DOI] [PubMed] [Google Scholar]
- Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- Musto H, Naya H, Zavala A, Romero H, Alvarez-Valin F, Bernardi G. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun. 2006;347:1–3. doi: 10.1016/j.bbrc.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kaneko T, Sato S, Mimuro M, Miyashita H, Tsuchiya T, et al. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- Nikodem P, Hecht V, Schlomann M, Pieper DH. New bacterial pathway for 4- and 5-chlorosalicylate degradation via 4-chlorocatechol and maleylacetate in Pseudomonas sp. strain MT1. J Bacteriol. 2003;185:6790–6800. doi: 10.1128/JB.185.23.6790-6800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H. Evolution of genome base composition and genome size in bacteria. Front Microbiol. 2012;3:420. doi: 10.3389/fmicb.2012.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H. Comparative analyses of base compositions, DNA sizes, and dinucleotide frequency profiles in archaeal and bacterial chromosomes and plasmids. Int J Evol Biol. 2012;2012:342482. doi: 10.1155/2012/342482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi WD, Edgcomb VP, Christman GD, Biddle JF. Gene expression in the deep biosphere. Nature. 2013;499:205–208. doi: 10.1038/nature12230. [DOI] [PubMed] [Google Scholar]
- Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, et al. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati A, Labutti K, Pukall R, Nolan M, Glavina Del Rio T, Tice H, et al. Complete genome sequence of Sphaerobacter thermophilus type strain (S 6022) Stand Genomic Sci. 2010;2:49–56. doi: 10.4056/sigs.601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A, Budin M, Brocks JJ. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA. 2003;100:15352–15357. doi: 10.1073/pnas.2536559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phale PS, Basu A, Majhi PD, Deveryshetty J, Vamsee-Krishna C, Shrivastava R. Metabolic diversity in bacterial degradation of aromatic compounds. OMICS. 2007;11:252–279. doi: 10.1089/omi.2007.0004. [DOI] [PubMed] [Google Scholar]
- Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, Whitman WB, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature. 2011;473:208–211. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- Roulis E, Polkinghorne A, Timms P. Chlamydia pneumoniae: modern insights into an ancient pathogen. Trends Microbiol. 2012;21:120–8s. doi: 10.1016/j.tim.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae) Environ Microbiol. 2012;15:1956–1968. doi: 10.1111/1462-2920.12001. [DOI] [PubMed] [Google Scholar]
- SÁNchez-Baracaldo P, Hayes PK, Blank CE. Morphological and habitat evolution in the Cyanobacteria using a compartmentalization approach. Geobiology. 2005;3:145–165. [Google Scholar]
- Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-Garcia C, et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA. 2012;109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass AM, Schmerk C, Agnoli K, Norville PJ, Eberl L, Valvano MA, et al. The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. ISME J. 2013;7:1568–1581. doi: 10.1038/ismej.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BJ, David LA, Friedman J, Alm EJ. Looking for Darwin's footprints in the microbial world. Trends Microbiol. 2009;17:196–204. doi: 10.1016/j.tim.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Shi T, Falkowski PG. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc Natl Acad Sci USA. 2008;105:2510–2515. doi: 10.1073/pnas.0711165105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Salem H, Kost C, Kaltenpoth M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae) Mol Ecol. 2012;21:6134–6151. doi: 10.1111/mec.12027. [DOI] [PubMed] [Google Scholar]
- Takaichi S, Maoka T, Takasaki K, Hanada S. Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2′-dirhamnoside. Microbiology. 2010;156:757–763. doi: 10.1099/mic.0.034249-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Yamashita A, Ishikawa J, Shimada M, Watsuji TO, Morimura K, et al. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 2004;32:4937–4944. doi: 10.1093/nar/gkh830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran K, Dollhopf ME, Aller R, Stackebrandt E, Nealson KH. Shewanella amazonensis sp. nov., a novel metal-reducing facultative anaerobe from Amazonian shelf muds. Int J Syst Bacteriol. 1998;48 (Pt 3:965–972. doi: 10.1099/00207713-48-3-965. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek. 2007;91:351–372. doi: 10.1007/s10482-006-9122-6. [DOI] [PubMed] [Google Scholar]
- Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, van Elsas JD, et al. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Micro. 2009;7:252–252. [Google Scholar]
- Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dyé F, Borziak K, Khalsa-Moyers G, Alexandre G, Sukharnikov LO, Wuichet K, et al. Azospirillum Genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet. 2011;7:e1002430. doi: 10.1371/journal.pgen.1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Raymond J, Wu M, Chatterji S, Ren Q, Graham JE, et al. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS One. 2009;4:e4207. doi: 10.1371/journal.pone.0004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Z, Hu S, Yu J. On the molecular mechanism of GC content variation among eubacterial genomes. Biol Direct. 2012;7:2. doi: 10.1186/1745-6150-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, Bokhorst S, Kang S, Zhou J, Greer CW, Aerts R, et al. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J. 2012;6:692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DC, Liu HC, Zhou YG, Schinner F, Margesin R. Tistrella bauzanensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2011;61:2227–2230. doi: 10.1099/ijs.0.026930-0. [DOI] [PubMed] [Google Scholar]
- Zhou J, Huang Y, Mo M. Phylogenetic analysis on the soil bacteria distributed in karst forest. Braz J Microbiol. 2009;40:827–837. doi: 10.1590/S1517-838220090004000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.