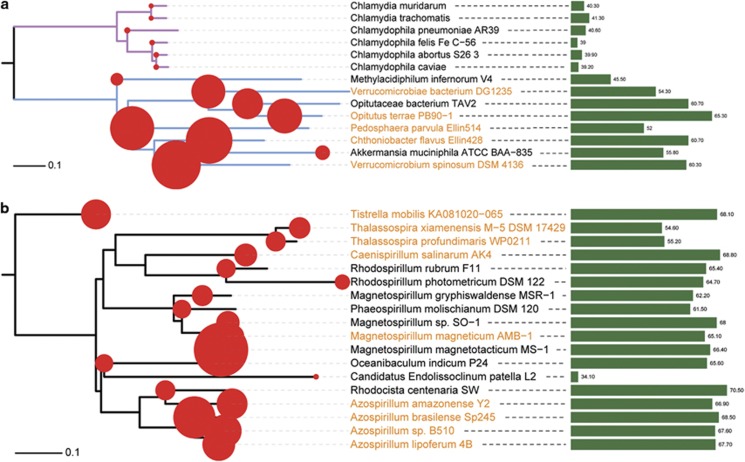
Figure 4.
The gain and loss of dnaE2 and its correlation with bacterial land colonization. Bacteria in Chlamydiae-Verrucomicrobia (a, with branches colored in magenta and light blue) and Rhodospirillaceae (b) are used for this case study. The phylogenetic trees are constructed by using MEGA 5 under JTT+Γ4 model (with 500 bootstraps). Red circles mapped on the trees are proportional to the genome size of each bacterium. Bacteria names labeled in brown color stand for DnaE2-containing bacteria and the green bars in the right panel are proportional to the GC content of each bacterium. Although the non-Azospirillum bacteria are correlated with aquatic environment, most of them actually dwell in the boundaries or mixtures of soil and water, such as water sediment (for example, M. magnetotacticum, M. gryphiswaldense and T. profundimaris) and ditch mud (for example, R. rubrum and P. molischianum).