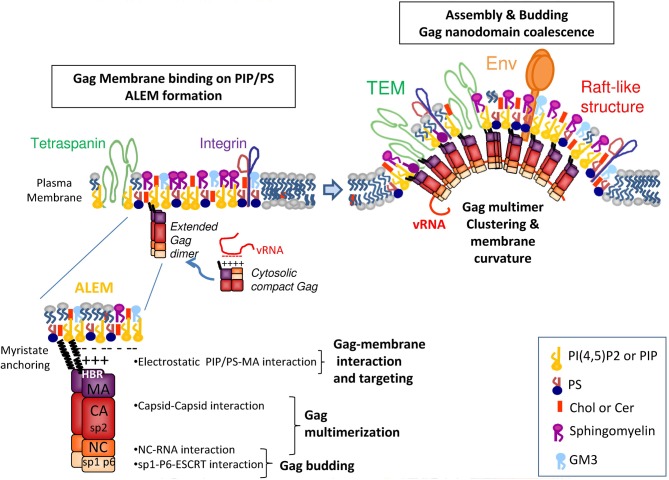
Figure 1.
HIV Gag assembly at the molecular level. The viral HIV-1 Gag protein is composed of several domains: the matrix MA is myristyolated in its N-term and contains a highly basic region (HBR) that binds the lipidic membrane or the viral genomic RNA, the capsid CA that favors Gag–Gag multimerization, the NC domain that selects the viral genome and favors Gag multimerization, its C-term p6 domain that binds ESCRT proteins requires for retroviral budding and particle release and 2 spacer peptides, sp2 and sp1 required for HIV assembly and morphogenesis. In the cytosol, Gag is a dimer and adopts a compact shape in which the viral genomic RNA is in interaction with the NC and MA domains. At the cell membrane, the MA domain of Gag targets, via its HBR, the inositol head of the PI(4,5)P2 located at the inner leaflet of the PM and its myristate anchors the lipid bilayer. The MA domain of Gag can recruit several PI(4,5)P2, and PS, thanks to its basic residues. This should allow the recruitment of other Gag molecules due to Gag electrostatic membrane interaction concomitant with Gag multimerisation, via CA, and NC on the viral RNA. This process should trigger the formation of ALEM (acidic lipid enriched microdomains) in the cell membrane. These ALEM containing Gag could now join together to create higher ordered microdomains of Gag and/or microdomains containing other proteins such as Env, or tetraspanins, integrins or other cell factors found in the viral envelope. The coalescence of these microdomains containing several Gag multimers creates the HIV assembly platform.