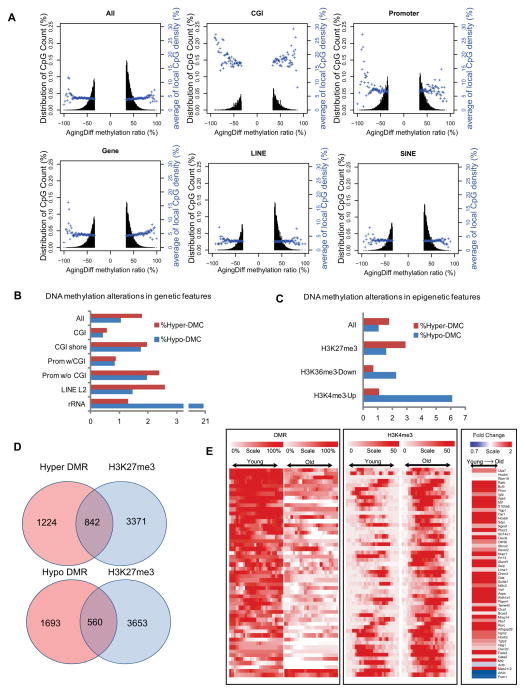
Figure 5. HSC methylome alterations with aging.
(A) Plots show the degree of differential methylation in young versus old HSCs and the relationship between the local CpG density (blue). The top left plot shows all DMCs in old HSCs defined as CpGs that are ≤ 20% less methylated and ≥20% more methylated. The other plots show DMCs located within different genomic features.
(B) The percentage of hyper- (red) or hypo- (blue) DMCs in different genomic features.
(C) Interaction of DNA methylation and histone modifications. The percentage of DMCs in the indicated epigenetic features and their alterations with aging.
(D) Venn diagram showing the overlap of DMR genes with polycomb repressed genes in HSC.
(E) Heatmap of DNA methylation, H3K4me3 signal and expression values for 50 potential HSC aging markers. Each row represents a gene-associated region where there is coexistence of differential H3K4me3 and DNA methylation. For H3K4me3 and DNA methylation, the red color denotes high methylation and white denotes no methylation. For gene expression, red denotes up-regulation and white denotes down-regulation.
See also Figure S5