Figure 7. DNA methylation regulates HSC function and contributes to age-related diseases.
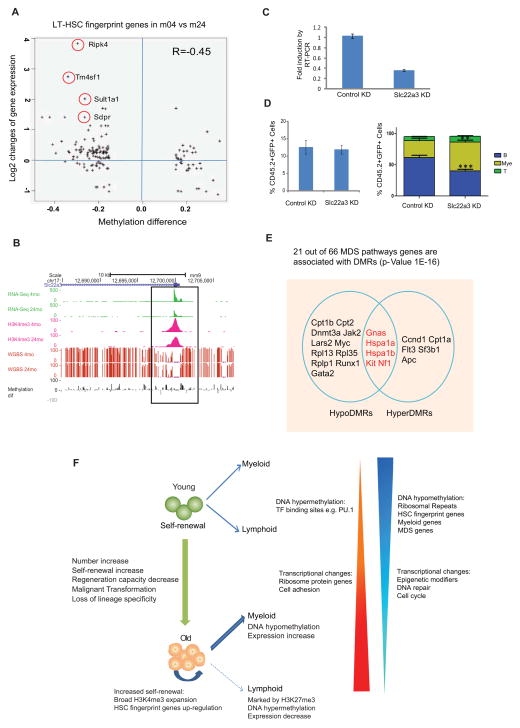
(A) The correlation between gene expression changes and methylation alterations for HSC fingerprint genes with DMR. The X-axis denotes the DMR methylation ratio difference, and the Y-axis denotes the log2 fold change of FPKM value for the gene. The circles mark the gene with names aside.
(B) UCSC browser track showing expression (green), DNA methylation (red bars), grey and black bars show differential methylation between 4m and 24m HSCs, H3K4me3 signal (pink) for the TSS region of Slc22a3 in young and old HSCs.
(C) QRT-PCR to show that Slc22a3 was knocked down by miRNA construct. Sca-1+ cells were enriched from 5-FU injected WT mice (4 month old) and transduced with retroviral miRNA construct. After in vitro culture for 2 days, 20,000 GFP+ cells were sorted for qRT-PCR.
(D) Left figure showed the contribution of retrovirally-transduced donor HSCs (CD45.2+GFP+) to recipient mouse peripheral blood. Right figure showed the percentage of indicated lineages within the retrovirally-transduced donor HSC-derived (CD45.2+GFP+) cell compartment in peripheral blood post-transplant (8-weeks). Myeloid cells (Mye) were defined using the markers Gr1+ and Mac1+, B-cells (B) are B220+, T-cells (T) are CD4+ and CD8. n=8 for each group. *** P<0.001, ** P< 0.01. Error bars represent Mean ± SEM.
(E) Venn diagram showing MDS genes with hyper or hypo-DMRs. Regions with overlap have both hyper- and hypo-DMRs within same gene (distinct regions).
(F) Model for functional and epigenetic alterations with HSC aging. DNA hypomethylation in ribosomal repeats could initiate chromosomal instability and indicate HSC activation state. DNA hyper- or hypo-methylation in master transcription factor binding sites could contribute to reinforcement of self-renewal program and inhibition of differentiation.