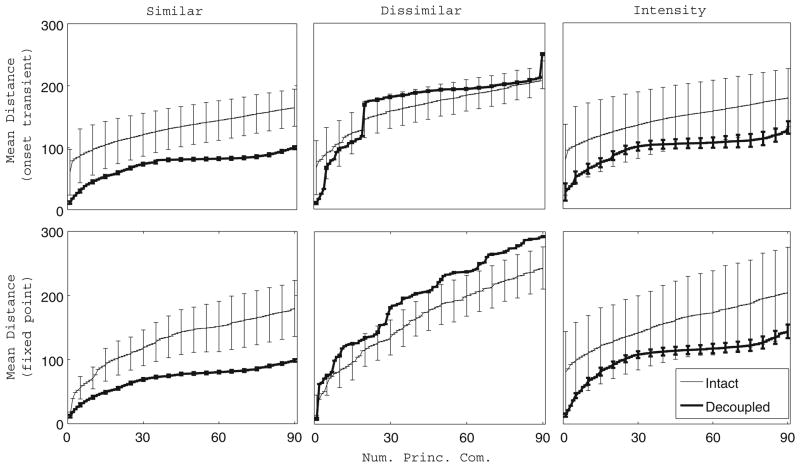
Fig. 8.
Distance between the principal component trajectories of two odors. Time-averaged Euclidean distance between the principal component trajectories of two odors plotted as a function of the number of principal components used in the computation (50 ms time bins). For a given (i,j) odor pair, the time-averaged intertrajectory distance during the onset transient (top row, 1–2 s) or fixed point (bottom row, 2.5–3.5 s) was computed between each trial of odor i and each trial of odor j (20 trials per odor). The plots show the time-averaged distance between a trial of odor i and a trial of odor j averaged over all possible trial-trial pairs, and the error bars represent the standard deviation of the trial-trial distance. The analysis was performed for two similar combinatorially coded odors (left column), two dissimilar combinatorially coded odors (middle column), and two intensity coded odors (right column) in the case of the intact network as well as the completely decoupled network. Trajectories of trials from each odor pair were projected onto the same principal components to enable computation of intertrajectory distances