Figure 3.
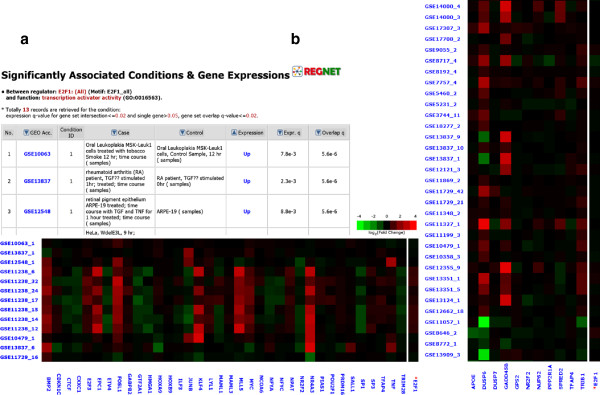
Condition table and heat maps for transcription networks of E2F1. (a) The condition table and heat map for the association between E2F1 and the gene set ‘transcription activator activity (GO:0016563)’. Several targets exhibit clear activations across a number of virus treated HeLa cells. Seven genes, Bmp2, E2f3, Epc1, Fosl1, Mll5, Myc and Nr4a3 are chosen for validation using a ChIP assay. (b) The heat map for the association between E2F1 and the gene set ‘negative regulation of protein kinase activity (GO:0006469)’. A part of the 55 conditions that associate E2F1 and the GO gene set are shown. Among the targets, Gadd45b and Dusp6 exhibit clear expression changes across a variety of microarray conditions. Both genes are tested using a ChIP assay.
