Figure 1.
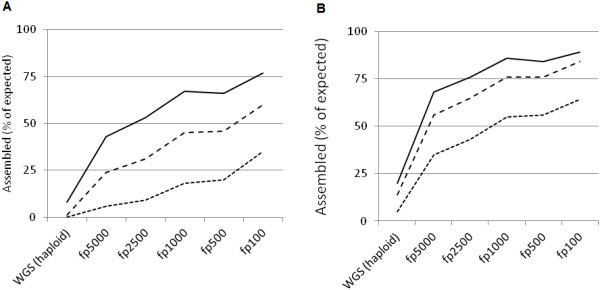
Assembly length achieved as a function of minimal contig size (bp) and pool complexity (number of Fosmids per pool). A: FP assembly alone. B: assembly with scaffolding. The leftmost point on each x-axis indicates the values achieved by haploid whole genome shotgun (WGS) sequencing. Solid, dashed, dotted lines: data for contigs longer than 5, 10, and 20 Kbp, respectively. Read coverage after read quality control and filtering was around 38x for the haploid WGS, and then 65x, 115x, 150x, 160, and 200x for the pools from fp5000 to fp100, respectively. The libraries used for scaffolding, 650 bp paired end and 2.4 kbp mate pairs, provided coverage around 20x and 25x, respectively.
