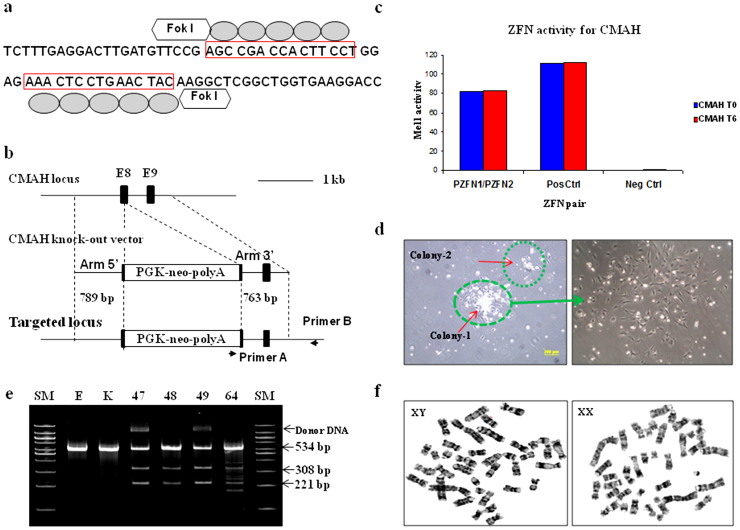
Figure 1. Efficient targeting of CMAH locus by ZFNs.
(a, b) Strategy to disrupt CMAH gene expression. Diagram of (a) illustrates a part of exon 8 of the CMAH locus and the sequence recognized by the ZFN pairs. The ZFN target sequences are boxed. Each ZFN consisted of a five finger protein that recognized the target sequence (gray oval) and the nonspecific cleavage domain of the Fok I restriction enzyme. (b) The homologous sequence used to promote homologous recombination (arm 5′ and 3′). The middle diagram illustrates the donor DNA used to disrupt CMAH expression. A neo expression cassette (PGK-neo pA) has been introduced between the homologous arms to allow antibiotic selection. Lower diagram shows the edited CMAH locus after homologous recombination with the donor DNA. (c) ZFN with confirmed activity in a yeast MEL-1 assay. (d) A single cell selection after ZFN-alone or ZFN + donor DNA transfection. (e) Surveyor assay (Cel-I) nuclease assay for ZFN-induced mutations in CMAH gene. Cel-I endonuclease digest of the 534 bp heteroduplex DNA derived from the hybridization of the DNA from control and each targeted cells into 308 bp and 221 bp fragments proofed the mutagenesis of one CMAH allele (line 48 and 64) or bialleles (line 47 and 49) in the targeted cells. Arrowheads indicate the expected positions of the digested products. SM, E, and K indicate size marker, control ear- and kidney-DNA amplified products, respectively. (f) Chromosome analysis in ZFN-targeted female (left) and male (right) somatic cells, which used for SCNT.