Summary
Ultra-high throughput sequencing of cDNA (RNA-Seq) is an invaluable resource for investigating alternative splicing in an organism. Alternative splicing is a form of post-transcriptional regulation in which primary RNA transcripts from a single gene can be spliced in multiple ways leading to different RNA and protein products. In plants and other species, it has been shown that many genes involved in circadian regulation are alternatively spliced. As new RNA-Seq data sets become available, these data will lead to new insights into links between regulation RNA splicing and the circadian system. Analyzing RNA-Seq data sets requires software tools that can display RNA-Seq read alignments alongside gene models, enabling assessment of how treatments or developmental stages affect splicing patterns and production of novel variants. The Integrated Genome Browser software program (IGB) is a free and flexible desktop tool that enables discovery and quantification of alternative splicing. In this protocol, we use IGB and a cold-stress RNA-Seq data set to examine alternative splicing of Arabidopsis thaliana LHY, a circadian clock regulator. Integrated Genome Browser is freely available from http://www.bioviz.org.
Keywords: genome browser, visualization, visual analytics, alternative splicing, A. thaliana, LHY, LHY1, circadian clock
1. Introduction
Most genes in higher eukaryotes contain introns, sequence segments that are removed from the primary RNA transcript either co- or post-transcriptionally. The process of removing introns, called splicing, involves stepwise assembly of a macromolecular complex called the spliceosome onto the nascent primary RNA transcript. The spliceosome catalyzes removal of the intron and ligation of flanking exonic sequences via two transesterification reactions involving the five prime and three prime splice sites of the intron, also called donor and acceptor sites, and a branch point adenosine residue upstream of the three prime splice site. The spliceosome consists of five ribonucleoproteins (U1, U2, U5, and U4/6 snRNPs) whose activity and interactions are modulated by a host of accessory proteins, including RNA helicases, SR proteins, hnRNP proteins, and others. Differential expression and activity of these accessory proteins influence splice site selection and also allow for different sites to be selected, a phenomenon known as alternative splicing. Alternative splicing (AS) can lead to production of alternative isoforms with distinct and sometimes antagonistic activities. Changes in the mRNA through differential inclusion of sequences can alter transcript localization, transcript longevity, or protein sequences. In animals, AS has been recruited as a regulatory mechanism enabling cellular diversity and differentiation, such as sex determination in insects and neuronal cell differentiation in the mammalian brain.
According to conservative estimates based on annotated gene models (1), around 20% of multi-exon genes in higher plants are thought to produce multiple transcript isoforms through AS. However, prior to the development of high-throughput sequencing of cDNA (RNA-Seq), it was difficult to estimate the prevalence and thus the biological significance of AS in plants. Studies that attempted to address the degree to which AS occurs in plants mainly used ESTs or tiling arrays to identify and quantify AS (2-4). Our analysis of Arabidopsis thaliana ESTs found that for most genes annotated as producing multiple variants, one isoform tended to predominate, and the number of ESTs supporting the minor form was typically less than one in ten (1). This result suggested that although many genes are capable of producing multiple isoforms through AS, the frequency with which this occurs is low. However, the study was limited by the heterogeneity of EST libraries as well as relatively small number of Arabidopsis ESTs (around 1.5 million) that were available at the time. We later repeated the analysis using new RNA-Seq data sets from Arabidopsis pollen and seedling and found essentially the same result: although around 15 to 20% of alternatively spliced genes were found to produce the less abundant isoform in significant amounts, most annotated AS events were rarely observed (5). Although this later RNA-Seq-based study involved many more sequences than the EST study that preceded it and thus had greater power to detect AS events, it should nonetheless be considered preliminary as only three libraries were sequenced. There will no doubt be many more RNA-Seq data sets published in future that will yield more information about AS, including its prevalence and function in plant species.
To help researchers take advantage of existing and future RNA-Seq data sets, we have added new features to the Integrated Genome Browser (6) that enable visual analysis of splicing patterns embedded in RNA-Seq data. This protocol explains how to use IGB to perform visual analysis of AS using Arabidopsis LHY as an example. LHY encodes a MYB transcription factor that together with CCA1 drives the morning loop of the circadian oscillator, a network of transcriptional regulators that activate or repress gene expression according to time of day. The LHY locus, similar to several other clock genes, undergoes extensive alternative splicing (for review, see (7)). LHY has been annotated by the Arabidopsis Information Resource (TAIR) as producing five distinct alternative splicing variants arising from alternative splicing in the 5′ UTR and from an exon skipping event affecting the coding region. Previous analyses of RNA-Seq data observed that LHY also produces splicing variants in which introns 4 and 9 are sometimes retained (8, 9). However, the relatively short read lengths of this early data set may have resulted in some AS events being missed. Another study that used a high-throughput qPCR panel to assess AS in circadian clock genes found additional novel splicing events in LHY, including an alternative acceptor site affecting intron 8 (10). By re-examining LHY using RNA-Seq data with longer read lengths and by examining this data in IGB, we can recapitulate previous findings, as well as report new aspects of LHY alternative splicing.
2. Materials
The RNA-Seq data used in this protocols paper were from two libraries prepared from cold-treated and control Arabidopsis thaliana seedlings. The data have not been published before now, and so we describe in detail how they were generated. Plants used in the experiment were sown onto soil in 4 inch pots and incubated for seven days in a Percival incubator set to 22°C, 45% relative humidity, under long day (16h/8h light/dark) illumination.. At zt4 (zeitgeber time, 4 hrs after lights on) on the seventh day, pots selected at random to undergo a cold stress treatment were transferred to a similarly configured Percival incubator set to 4°C. Relative humidity (RH) was adjusted to 75% for each incubator as the colder incubator RH was difficult to maintain below this level. Following the transfer, nine samples from control and cold-treated samples were collected 45 min later,, at zt7, zt10 and zt16 on the first day of treatment; zt7 on the second day; zt4, zt11 and zt17 on the third day; and zt2 on the fourth day. The above ground parts (shoots) were collected from two pots per treatment at each time point. Samples were flash-frozen on liquid nitrogen and stored at −80°C prior to RNA extraction. Frozen samples from all time points were pooled, RNA was extracted, and cDNA libraries were prepared for Illumina sequencing as described previously (5), keeping treatment and control samples separate. Each library (treatment and control) was sequenced on an Illumina GAIIx sequencer for 75 cycles on two lanes per library, generating more than 40 million reads per library. The resulting 75 base, single-end sequences were aligned onto the latest Arabidopsis genome assembly (TAIR9, released in June 2009) using the TopHat spliced alignment tool (11).
Reads were then sub-divided into two groups: reads that mapped to one location in the genome (SM, or single-mapping reads) and reads that mapped to more than one location in the genome (MM, or multi-mapping reads). The protocol and images described here refer to the SM reads only. The version of IGB used in this protocol is a pre-release version of IGB 7.1 corresponding to subversion repository revision 15,286. Alignment files are available from the main IGBQuickLoad site (igbquickload.org) and reads are available from the Short Read Archive via accession SRP029896.
3. Method
3.1 Start IGB and open data sets
Go to http://www.bioviz.org and follow the links labeled “Download” to download and start IGB. IGB can run on any computer that has Java installed, but methods of launching IGB may vary depending on your system. For discussion of how to launch to IGB, see Note 1.
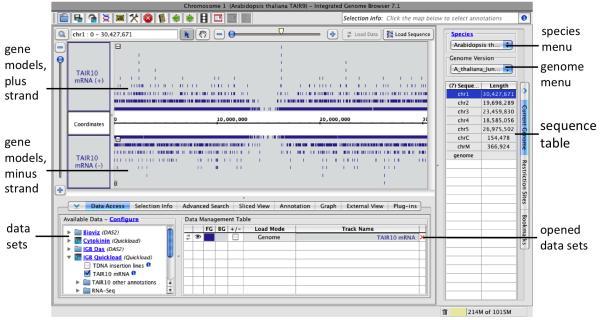
Open the latest Arabidopsis genome and reference gene models by clicking on the A. thaliana image on the start screen. Clicking the image triggers loading of the latest Arabidopsis genome assembly and associated gene models from the IGBQuickLoad.org Web site directly into IGB (Figure 1). The data loading process may take several seconds or a few minutes depending on your network connection.
Open cold stress and control RNA-Seq data sets. In the Available Data panel on the left side of the Data Access tab, open folder IGB Quickload > RNA-Seq > Loraine Lab > Mixed Cold > SM > Reads and select Control, alignments and Cold, alignments. Position the mouse over the sample labels to view a tooltip describing the data sets or click the “i” icon to open a Web page with details about the data sets. When you select a data set or open a file, the data set appears in the Data Management Table and a placeholder track appears in the Main View. See Note 2 for discussion of opening files from your desktop or Web sites.
Go to the LHY gene by entering “LHY” in the search box at the top left of the display. Note that when you enter this search term, IGB suggests several options, including “LHY1/LHY.” (LHY1 is a synonym of LHY according to the TAIR10 annotations.) Use the up and down arrow keys or the mouse to select option “LHY1/LHY” and press ENTER to run the search. After the search, the Main View will zoom and pan to a close-up view of the LHY gene centered in the display.
Load the sequence data by clicking the Load Sequence button at the top right of the display to load the reference sequence, which appears as a gray bar in the Coordinate Axis track. Click within the gene to set the zoom focus and drag the horizontal zoom slider at the top of the display to the right to zoom in to view the sequence. You can also zoom by clicking the + button to the left of the slider, click-dragging a region in the coordinates axis, or double-click an annotation feature. As you zoom in, the individual bases become visible. To re-center the LHY gene in the display, use the horizontal zoom slider or double-click on an intron. See Note 3 for more details on how to zoom in IGB.
Load the alignments data by clicking the button labeled Load Data at the top right of the display (see Note 4). When you click Load Data, the two previously empty tracks are filled with stacks of reads whose alignments correspond to the intron/exon structure of the LHY gene.
Figure 1. Integrated Genome Browswer after selecting the Arabidopsis genome.
Plus and minus strand gene models from the TAIR10 mRNA gene model annotations are shown.
3.2 Adjust Main View
To facilitate visual analysis of splicing data, it is useful to configure the layout and presentation of reads and annotations in ways that reduce clutter and make biologically meaningful patterns easier to recognize. To adjust the view for splicing analysis of LHY, perform the following steps:
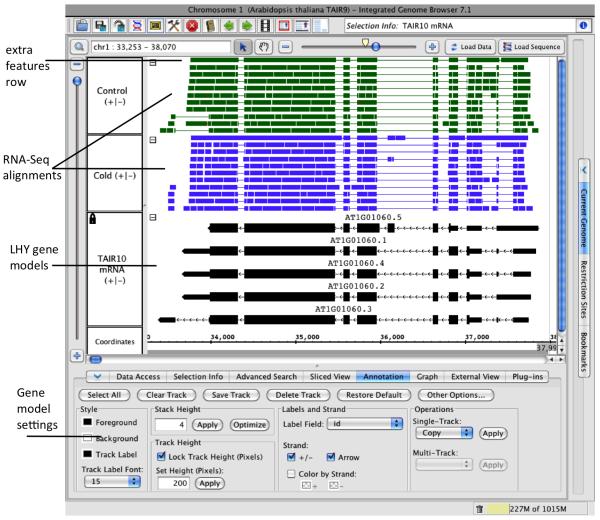
Edit track labels. When you first open a file from the IGBQuickLoad site, the full folder path to the file is displayed in the track label. To simplify the view, edit the track names. In the Data Management Table, click a track name to edit the track name. After editing, press ENTER to trigger the change in the Main View.
Combine plus and minus gene model tracks and adjust stack height. Click the track label to the left of the TAIR10 mRNA track to select it. Select the Annotation tab at the bottom of the IGB window and choose the option labeled +/− under the Strand section to combine plus and minus strands into a single track. Also, enter “4” in the stack height section. This will cause IGB to use less vertical space to display the LHY gene models, which will be useful when other data sets and tracks are loaded. See Note 5 for discussion of strand options and stack height.
Make more space for visualization. Expand the space on your desktop available for visualization by hiding the Current Genome or other tabbed panels and enlarging the IGB window. After enlarging the IGB window, re-position the divider separating the main view and the bottom tabs so that the tabs occupy smallest are possible while still keeping the controls visible.
Lock track height. To ensure that the LHY gene models will remain the same size, no matter how much data is loaded into the main view, lock the gene model annotations track height. To lock track height to 200 pixels, select the track by clicking the TAIR10 mRNA track label, select the Annotation tab, choose Lock Track Height (Pixels), and enter 200.
Modify foreground, background, and track label colors. Select tracks by clicking the track labels and use the Style section of the Annotation tab to change the color scheme for tracks. Also use the font option to increase or decrease the track label font size. To match figures shown in this article, use the following color scheme: white background for all tracks, black track labels for all tracks, black foreground for the reference gene models track (TAIR10 mRNA), and green and blue foreground colors for the control and cold stress alignment/read tracks, respectively.
Modify edge-matching color. For visual analysis of splicing, IGB provides an edge matching function designed to facilitate rapid comparisons between gene models and other items in the display. To activate edge matching, click the label of a gene model to select it and observe that the edges of all other items in the display that have matching boundaries become highlighted. To make these highlights easier to see with our chosen color scheme, choose File > Preferences > Other Options and change the Edge Matching color from white to red or black. See Note 6 for discussion of edge matching.
Clamp to view. When viewing a single gene in detail, it is useful to restrict zooming to that one gene of interest. To restrict zooming to the LHY region, zoom to center LHY in the center of the display and select View > Clamp to View. While Clamp to View is active, the range of zooming and panning will be limited to the LHY region. After performing the preceding adjustments, the IGB window should look like the image shown in Figure 2.
Save your work. After loading the data and configuring the IGB display, use the IGB session saving feature to save your work as an IGB session file. The next time you use IGB, you can load the IGB session file and begin again from this point, with colors, files and other choices remembered and loaded. To save the session, choose File > Save Session. Note that Load Session, which you would use to open an IGB session file after re-starting IGB, is also in this menu.
Figure 2. Adjusting the Main View to Optimize the Data Analysis Environment.
The region surrounding the LHY gene is shown. The display has been adjusted as described in the section 3.2.
3.3 Create and analyze junction tracks
Genomic alignments of RNA-Seq reads derived from exon-exon junctions in spliced mRNAs typically contain gaps. These gaps define the former locations of introns and can be used to identify and analyze AS. Within the IGB display, gapped reads appear as alignment blocks separated by thin lines corresponding to the gaps. To facilitate analysis of splicing using gapped reads, IGB provides a visual analytics function called “FindJunctions” that you can use to quantify splicing choices and assess the prevalence of AS within an RNA-Seq data set.
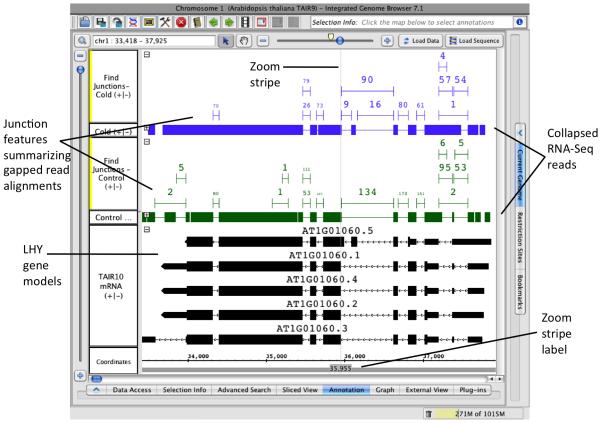
Create FindJunctions tracks for each RNA-Seq data set. To create junction tracks for each RNA-Seq data set, select all of the sample tracks (SHIFT+click the track labels), select Single-Track: Find Junctions in the Annotation tab, and click Apply. For each selected track, a new track will appear that contains junction features summarized from the read alignment tracks. Each junction feature has a numeric label indicating the number of gapped reads that supported the junction. By default, only single-mapping reads that align with at least five bases on either side of the intron are counted, and you can change these defaults by right-clicking a track and selecting the Track Operations > FindJunctions > Configure… menu item. See Note 7 on configuring FindJunctions.
Simplify the display by collapsing the alignment tracks. Click the - icon in the upper left corner of the alignment tracks to collapse the reads into a single row.
Re-organize the tracks. Click the track labels and drag the junction tracks above their corresponding read tracks as shown in Figure 3.
Use IGB zoom controls and edge matching to inspect the junctions and compare them to the annotated gene models. The FindJunctions track from the control sample contains reads 134 reads that support skipping of exon 5 (82 bases, 36,171 to 36,089) and no reads that support its inclusion. However, the cold stress sample contains 90 reads in support of exon skipping and 9 and 16 reads supporting exon inclusion. Consistent with a previous examination of alternative splicing in LHY (10), cold stress increased the prevalence of the exon inclusion in LHY. Note that when you select a junction read, its name appears in the Selection Info box at the top right of the IGB display and the name indicates the location of the intron inferred from the gapped reads used to create the junction. Make note of junctions that have no corresponding intron in the LHY gene models. Some of these have only one or two supporting reads, while others have several. Among the best-supported novel introns with respect to the annotated gene models is junction feature J:chr1:33858-33980, which has five supporting reads and indicates a new splicing event affecting the three prime end of the transcript. Another novel junction is J:chr1:35963-36089, which is immediately to the left of the optionally included exon (see Figure 3) and thus is present in the cold stress and not the control data set. The donor site associated with this junction does not match any of the annotated gene models but the acceptor site matches the boundary of the optionally included exon, as described in (10). The RNA-Seq alignments data suggests that the acceptor site corresponding to this novel intron is the only one that is used and that the annotated acceptor is incorrect.
View the effects on protein sequence. In IGB, translated regions are shown as tall blocks with shorter blocks representing the five and three prime untranslated regions (UTRs). If the sequence has been loaded (which you did in an earlier step), you can zoom in on a location and observe the amino acid translation of a gene model. Zoom in on the option (sometimes skipped) exon 5, and look at the amino acid sequence. Compare the translations of the different gene models and note that including exon 5 does not introduce a premature stop codon. See Note 8 for discussion of viewing sequence data in IGB.
Figure 3. Junction tracks created from RNA-Seq read tracks.
Junction tracks derived from read alignment tracks are shown above the read alignment tracks used to create them. To conserve vertical space, read tracks are collapsed. Numbers above each junction feature indicate the number of gapped reads that supported it. The zoom stripe is positioned close to the novel acceptor site inferred from junction J:chr1:35963-36089.
3.3 Use depth graphs to detect intron retention
Depth graphs, also called coverage graphs, summarize the number of reads that overlap each base position within a genome. Depth graphs are useful for identifying intron retention (IR), a form of AS that is especially common in plant species. Depth graphs can also provide a rough estimate of overall gene expression within a sample.
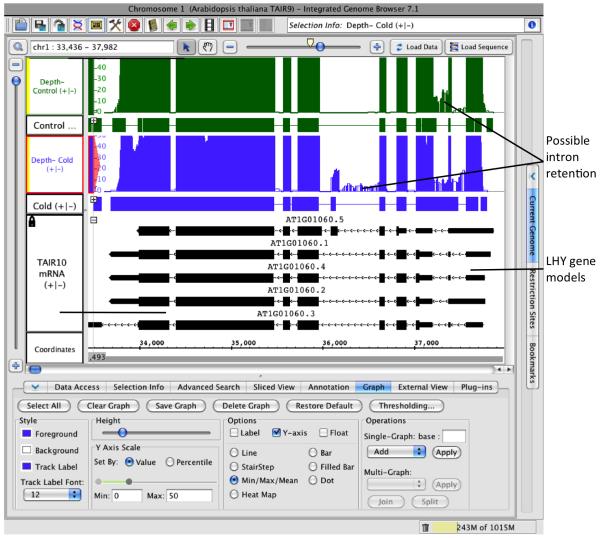
Create depth graphs. To create depth graphs for the RNA-Seq tracks and check for retained intron AS events, select the tracks as before (see preceding section), choose Single-Track: Depth in the Annotation tab, and click Apply. See Note 9 for discussion of coverage graphs. Use the Data Access tab as before if you wish to re-label the new depth graphs.
Simplify the display by hiding junction tracks. Since you will now focus on identifying introns, simplify the display by hiding tracks that are no longer needed. Right-click the track labels next to the junction tracks and select Hide or use the track visibility icon (eye) next to the track’s listing within the Data Access tab.
Adjust the scale of the depth graphs. To use the depth graphs to find IR AS events, decrease the scale so that lower coverage regions are more obvious. Click the Graph tab, click Select All button to select all graphs; in the section Y-axis Scale > Set by: Value, enter 20 in the Max text box to limit the upper boundary of the Y-axis. Observe that introns two and five have some coverage, indicating these introns may sometimes be subject to intron retention (Figure 4). See Note 9 for discussion of graph scaling.
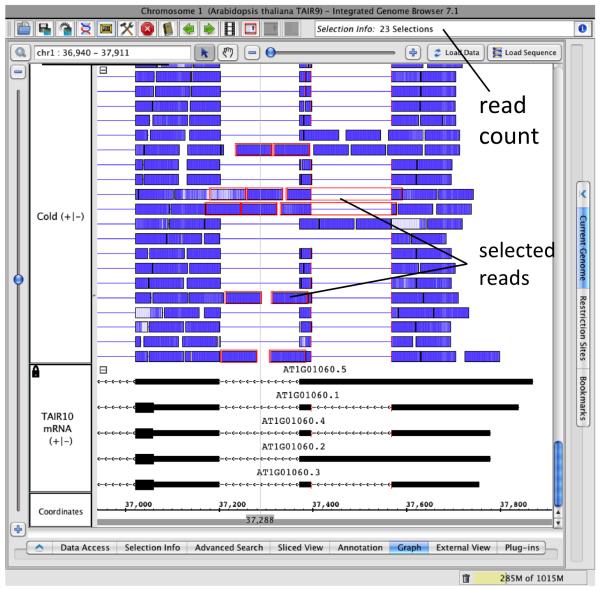
Count retained intron reads to assess frequency of intron retention. Hide all the tracks except the gene models, and cold reads. Adjust the horizontal zoom level to enlarge intron two so that it occupies the entire IGB screen and use View > Clamp to View to restrict zooming to this region. Expand the read track using the + icon, then select the track and click Optimize under the Annotation tab to show all the reads in the region. Use the vertical zoom controls to stretch the read track vertically so that you can see each read. Using the mouse, select all the reads that map within the intron. Use SHIFT-CLICK to select multiple reads and CNTRL-SHIFT-CLICK to remove reads from the current selection as needed. The Selection Info box (upper right) reports the number of items currently selected, which you can use to count the reads that overlap the intron (Figure 5). Although IR does appear to occur, the number of gapped reads outnumbers the retained intron reads, suggesting that mRNAs with retained introns probably do not account for a large proportion of LHY transcripts.
Figure 4. Coverage graphs created from RNA-Seq read tracks.
Coverage graph track derived from read alignment tracks are shown above the read alignment tracks used to create them. To conserve vertical space, read tracks are collapsed. Each graph track has a scale on the left that shows the number of reads overlapping positions indicated in the Coordinates track sequence axis.
Figure 5. Counting reads to quantify intron retention.
All tracks except the gene models (TAIR10 mRNA) and cold stress RNA-Seq read alignments track are hidden. The stack height for the RNA-Seq read alignment track is optimized to allow all reads to be shown and the vertical zoom has been used to stretch the display vertically. Reads that appear to support intron retention are selected and the number of selected reads is shown in the Selection Info box. Some of the selected reads are off-screen, above the current view.
4. Notes
Start the latest version of IGB by selecting a web-start option at http://www.bioviz.org. If your computer system does not support downloading applications via Java Web Start, then download and unzip the file labeled igb.zip available from the IGB download page. Unpacking the igb.zip file will create a new folder named “igb” on your computer. Open the igb folder and double-click a “.bat” IGB start script file (Windows) or “.command” start script (Mac) to start IGB. Note that the start scripts specify different amounts of memory that IGB will use when running, and the memory version you choose will vary depending on your computer. We recommend that if you have a Mac computer with 8 Gb or more of RAM, you can safely run a 5 Gb version. Windows users with less than 4 Gb of RAM should run the 1 Gb version.
In addition to loading data from the IGBQuickLoad.org site, you can also open data files stored on your local computer or Web sites using the File > Open File… or Open URL… menu options. In addition to using the File menu, you can also open a file by click-dragging it from your desktop or from a Web site into the IGB window.
IGB is unique among genome browsers in that it supports rapid, smooth, and flexible zooming. There are two types of zooming in IGB: animated zooming, in which the display appears to expand or contract in a animated fashion, and jump zooming, in which the display “jumps” to a new location. To zoom in IGB, use the horizontal or vertical sliders (animated zooming) or click one of the zoom buttons next to the sliders (jump zooming). When you zoom in IGB, the zoom stripe, the vertical line that marks the location of your last mouse click within the main IGB window, remains stationary while the rest of the view expands or contracts around it; use the zoom stripe to focus zooming and also as a pointer when comparing boundaries of overlapping items. To jump-zoom to an item in the display, double-click it. To zoom to an entire gene model (versus a single exon within a gene model), double-click an intron (thin line) or double-click the gene model’s label. To jump-zoom to a region, click-and-drag over a region in the coordinate axis. You can also enter coordinates in the region box in the upper left of the display or search for a gene by name to jump-zoom to a new location in the genome.
RNA-Seq data sets are large, comprising many millions of sequence read alignments. For this reason, IGB does not automatically load an entire RNA-Seq data set directly into the viewer but instead waits until you request data by clicking the Load Data button. Note that the Data Management Table within the Data Access tab reports a Load Mode setting for each track, indicating how the data will be loaded into IGB. The mRNA track load mode is Genome, which means the entire data set has been loaded into IGB, a reasonable setting for this data set, which contains 30,000 or fewer features. The load mode for the RNA-Seq data sets (cold and control) however, is set to Manual, meaning that data will only be loaded into IGB when you request it. This is because each RNA-Seq data set contains tens of millions of features, more data than IGB can display at once. Also note that when you click Load Data, only data for the currently visible region is loaded into IGB.
When IGB first loads reference gene model annotations or RNA-Seq reads, it reserves enough vertical space to display up to ten (the default) overlapping features in distinct rows within a track. If there are locations that contain more than ten items that overlap along the sequence coordinates axis, the additional models will be indicated in the top row of the track, the “extra features” row. However, when viewing a gene or region with fewer than ten gene models, you will observe some empty space above the models. You can eliminate this extra empty area by changing (or optimizing) the track’s stack height setting, which is the number of rows that can be shown per track in addition to the extra feature row at the top of the track. Also by default, IGB separates plus and minus strand gene models into separate tracks. However, when viewing a single gene, it often is better to combine the tracks for a more compact view by selecting the “+/−” combine strands setting. If you also select the “arrow” option, then gene models will be shown with arrowheads on the ends of the transcript and “greater” or “less” than symbols within introns to indicate the direction of transcription and strand of origin.
Edge matching is a visual analytics technique that was introduced by an earlier version of IGB called Neomorphic’s Annotation Station, which was used to annotate early assemblies of the Arabidopsis genome. Edge matching is designed to enable rapid comparisons between gene models and alternative splicing variants. To use edge matching, select a gene model or an exon within a gene model and observe that all other items with matching boundaries acquire an edge match highlight on compatible edges. However, the default edge match highlight color is white, which means that if you have changed your track background color to white, the edge matching highlight may be difficult to see. IGB allows you to change the edge match color by selecting File > Preferences > Other Options. Depending on your color scheme, red is usually a good choice for the edge match color setting.
The FindJunctions feature can be configured to consider single-mapping reads only or consider only reads with a threshold value of flanking bases on either side of the gap. The default is to use single-mapping reads that have at least five bases on either side of the gap. When junctions are created, the size of the blocks on either side of the gap is the same as the flanking threshold that was used in order to provide a visual cue indicating how the junctions features were created. However, if you choose “TopHat Junctions,” the size of the flanking blocks will match the size of the largest flanking blocks present in the original gapped reads.
Right-click a gene model and select View Genomic Sequence in Sequence Viewer to open a new window that displays the sequence of the selected item. The Sequence Viewer has options to show spliced (cDNA) and unspliced (genomic) sequence and protein translations in different frames. You can also copy and paste sequence from both the Sequence Viewer and from the genomic sequence displayed below the coordinates axis. Also, zooming in on a gene model within the Main View reveals its amino acid translation.
You can set the Y-axis scale of a graph either by Value or by Percentage. Value is an absolute number; percentage is the percentage of all results currently loaded. If you load several genes, one of which has a very high peak, but choose to focus on a lower abundance region, IGB will still calculate the by Percentage based on all the loaded data, so your Y-axis may need to be reduced more than you expected to show the full height of the region you are viewing. Alternatively use the by Value option and set it to a usable height.
References
- 1.English AC, Patel KS, Loraine AE. BMC Plant Biol. 2010;10:102. doi: 10.1186/1471-2229-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leviatan N, Alkan N, Leshkowitz D, Fluhr R. PLoS One. 2013;8:e66511. doi: 10.1371/journal.pone.0066511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Plant J. 2004;39:877–85. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 4.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. Nucleic Acids Res. 2004;32:5096–103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loraine AE, McCormick S, Estrada A, Patel K, Qin P. Plant Physiol. 2013;162:1092–109. doi: 10.1104/pp.112.211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicol JW, Helt GA, Blanchard SG, Jr., Raja A, Loraine AE. Bioinformatics. 2009;25:2730–1. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Santangelo S, Schlaen RG, Yanovsky MJ. Brief Funct Genomics. 2012;12:13–24. doi: 10.1093/bfgp/els052. [DOI] [PubMed] [Google Scholar]
- 8.Filichkin SA, Mockler TC. Biol Direct. 2012;7:20. doi: 10.1186/1745-6150-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JW, Nimmo HG. Plant Cell. 2012;24:961–81. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]