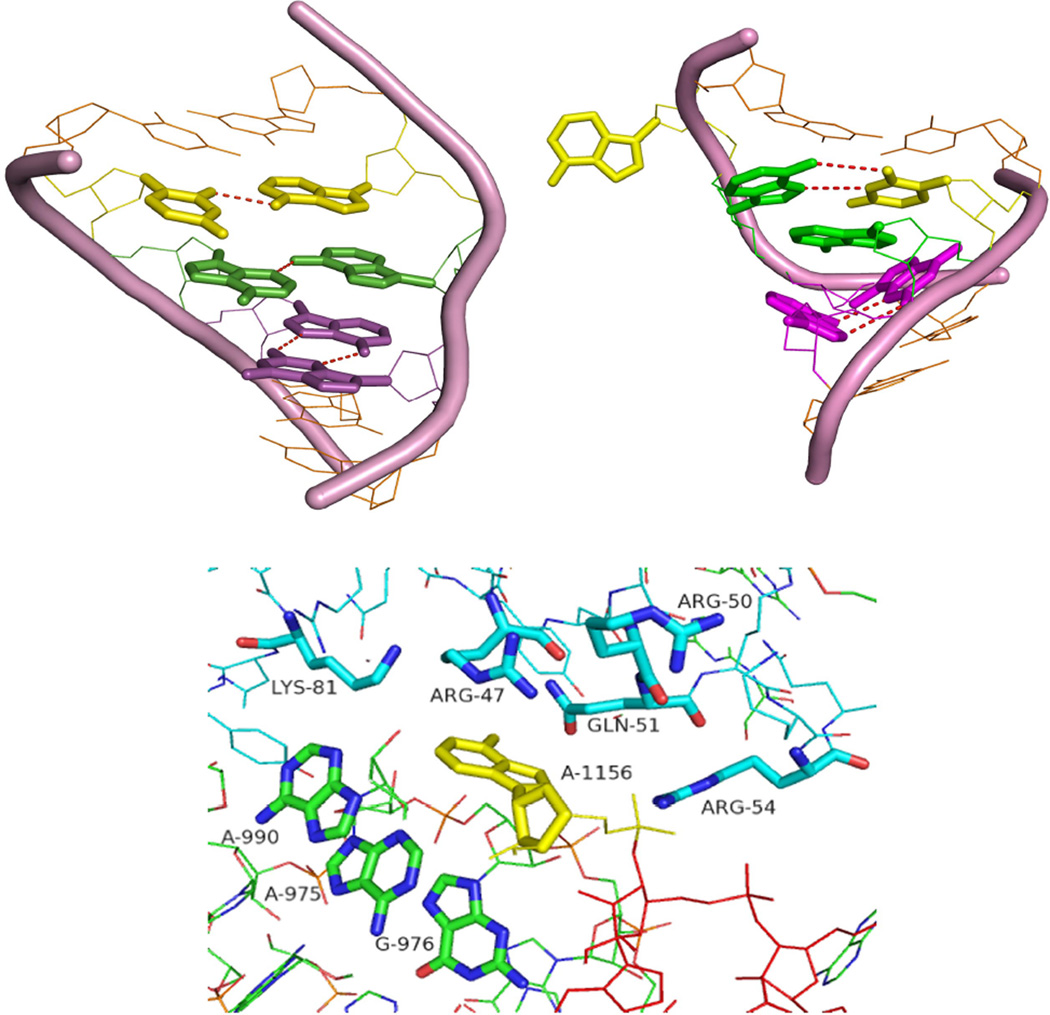
Figure 9.
A comparison between internal loop structures obtained by NMR (left upper panel) and crystallography of the D. radiodurans large ribosomal subunit (right upper panel) (4). Analogous base pairs are colored the same. The flanking pairs and the backbone of the loop are shown in orange. In the NMR structure, A15 and U4 (yellow) are paired in a cis Watson-Crick/sugar edge pair. In the crystal structure, A15 is bulged out and U4 forms a reverse Hoogsteen pair with A14. In the NMR structure, A5 and A14 (green) form a sheared pair, but A5 and A14 are stacked in the crystal structure. G13 and A6 (pink) form a sheared pair in both cases. Lower panel shows the tertiary contacts close to the bulged adenine (A1156) in the E. coli crystal structure (residues within 5 Å). The overall folding of the loop in E. coli is the same as for D. radiodurans (RMSD =0.67 Å)