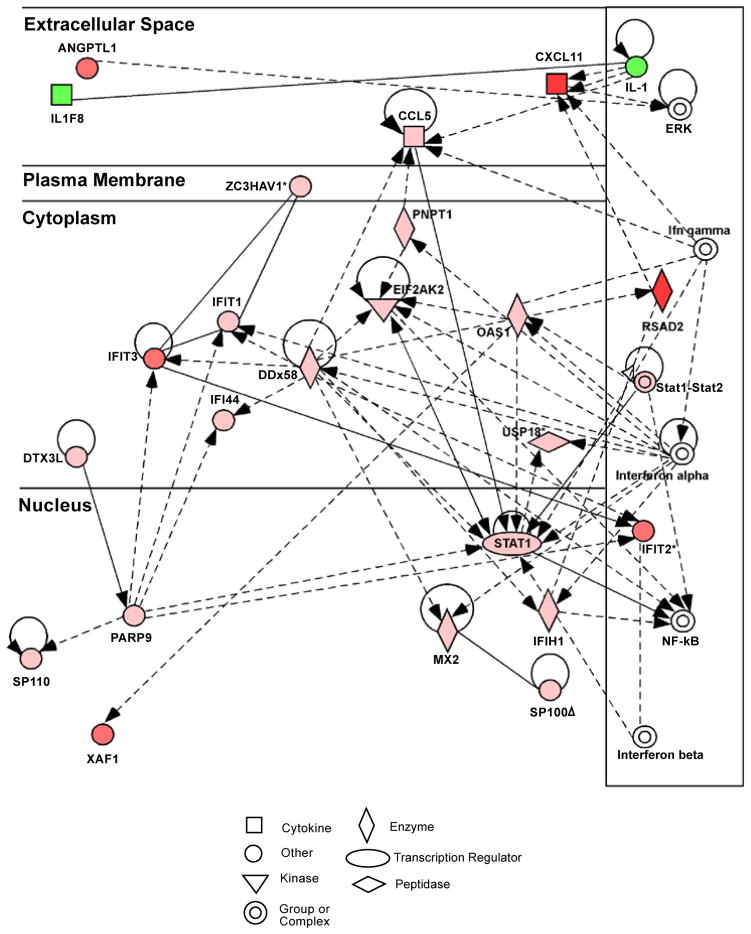
Figure 2.
Molecular network generated by Ingenuity Pathway Analysis (IPA) of highly significant gene changes in human RPE cells after in vitro stimulation with AGE (10 μg/mL). Colored symbols represent genes that were significantly highly upregulated (red) with decreasing relative levels indicated by lighter shades (pink and light pink) or downregulated (green) in our data set [11]. The white entries are molecules from the Ingenuity database, inserted to connect all relevant molecules in a single network. Solid lines indicate known direct physical relationships between molecules, while dashed lines indicate known indirect functional relationships. Note the chemokine, CXCL11, and RSAD2 (viperin) are shown to be highly upregulated in this network, and were also associated with drusen in postmortem donor eyes [12]. The top two functionalities identified by Ingenuity for this molecular network are “Interferon Signaling,” “Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses”.