Figure 4.
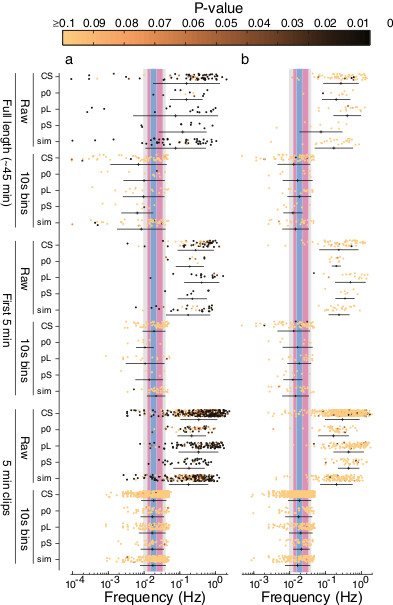
Lomb-Scargle periodogram analysis of multiple genotypes. Each datum represents the frequency with maximum power in a single song. The P-value for each value is pseudocolored according to the colorbar shown along the top of the figure. The mean ± 1 standard deviation is shown below each cloud of points. Each row contains data from a different genotype (D. melanogaster Canton-S (CS), period [Null] (p0), period [Long] (pL), period [Short] (pS), or D. simulans (sim)) analyzed in different ways. On the top, full-length (45 min) recordings were analyzed either as raw inter-pulse interval data or with data summarized as the means of 10 s bins. In the middle, the first 5 min after a male started singing were analyzed for each recording. This mimics most closely the analyses performed in most previous studies. On the bottom, full-length songs were divided into 5 min clips and each clip was analyzed separately. (a) The full-length raw data for all genotypes displayed significant rhythms across a wide frequency range on the order of seconds. These high-frequency signals are retained in the 5 min clips. Binning the data truncates the high frequency range that can be sampled, eliminating the statistically significant high frequency rhythms. The remaining maximum Lomb-Scargle periodogram peaks cluster near the previously reported range for KH cycles (illustrated with blue, red, and grey bands, as in Figure 3), although most of these values are not statistically significant. (b) Randomized song data display similar patterns as the real data, except that the high frequency rhythms in randomized data are mostly not statistically significant.
