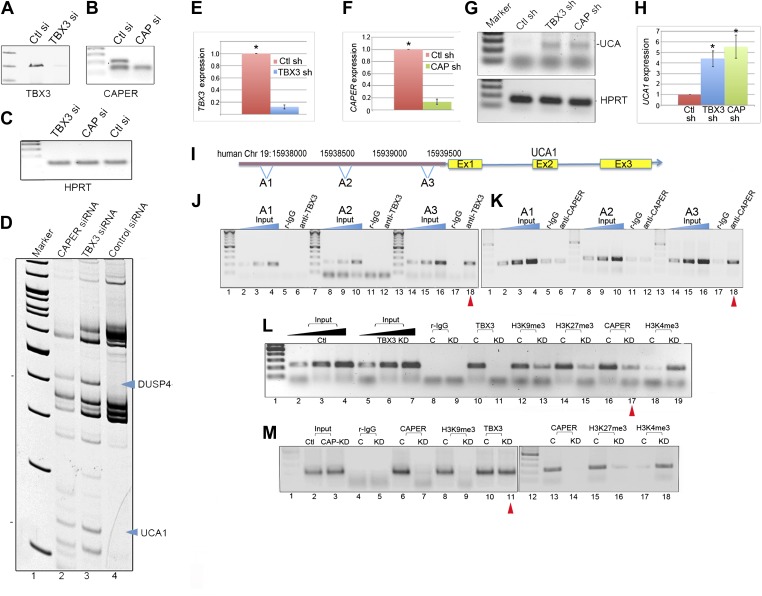
Figure 4. CAPERα/TBX3 directly represses expression of the long noncoding RNA UCA1.
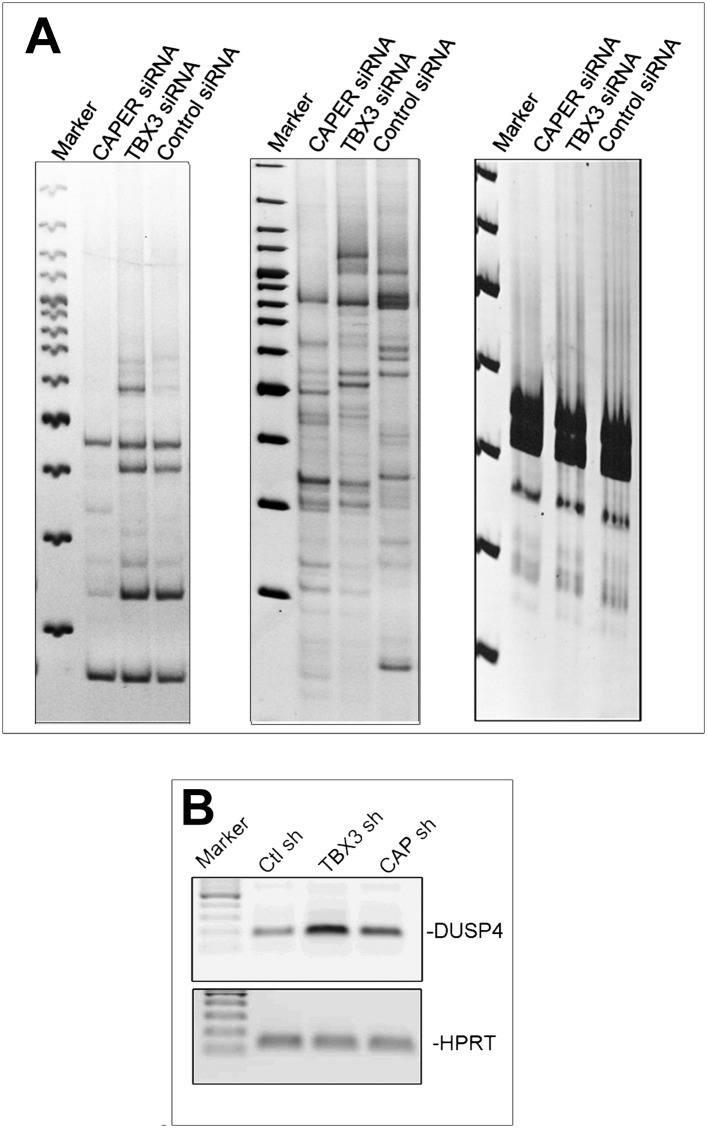
(A–C) Gel showing RT-PCR analysis of TBX3, CAPERα, and HPRT expression in control, TBX3 and CAPERα siRNA-transfected HEK293 cells. The siRNAs effectively decreased transcript levels of their targets. (D) Differential display: representative PAGE gel of cDNAs derived from random primed, RT-PCR'd mRNAs from CAPERα, TBX3 and control siRNA transfected HEK293 cells. Blue arrowheads denote upregulated transcripts subsequently identified by sequencing as DUSP4 and UCA1. (E and F) qPCR analysis of TBX3 and CAPERα transcript levels in control and TBX3 or CAPERα shRNA transduced HFFs (repeat of experiment shown in Figure 2—figure supplements 1A and 2A). (G) RT-PCR analysis of UCA1 and HPRT gene expression in control, TBX3 or CAPERα shRNA-transduced HFFs. (H) qPCR analysis of UCA1 transcript levels in control, TBX3 or CAPERα shRNA transduced HFFs. Results confirm differential display result that KD of TBX3 or CAPERα results in increase in UCA1 transcript levels. (I) Schematic representation of the UCA1 locus with primer sets employed for ChIP-PCR amplification of denoted regions 5′ of gene (A1, A2, A3). (J) Anti-TBX3 ChIP-PCR of regions of the UCA1 promoter in HFFs; only A3 is ChIP'd by TBX3 (lane 18, red arrowhead). (K) Anti-CAPERα ChIP-PCR of regions of the UCA1 promoter in HFFs; only A3 chromatin is ChIP'd (lane 18, red arrowhead). (L) ChIP-PCR analysis of UCA1/A3 chromatin from in HFFs transduced with control (C) or TBX3 (KD) shRNA; ChIP antibodies are listed at top. Note decreased CAPERα binding after TBX3 KD (lane 17, red arrowhead), gain of activating mark H3K4me3 and loss of repressive marks H3K9me3 and H3K27me3. (M) ChIP-PCR analysis of UCA1/A3 with antibodies listed at top of panel in HFFs transduced with control (C) or CAPERα shRNAs. Note continued TBX3 binding despite CAPERα KD (lane 11, red arrowhead) and changes in chromatin marks parallel those seen in with TBX3 KD in panel L. TBX3, CAPERα = human; Tbx3, Caperα = mouse.