Figure 3.
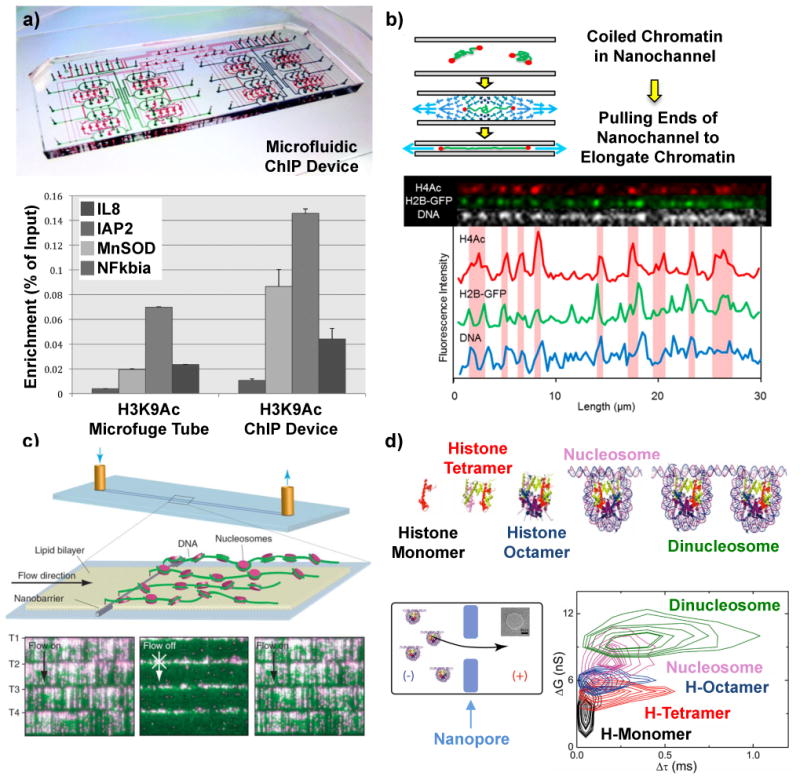
Micro and nano methods for mapping histone modifications and nucleosome arrays. a) Microfluidic device to perform automated chromatin immunoprecipitation (ChIP) reactions with higher efficiencies than conventional protocols (bottom panel)68. (b) Optical mapping of fluorescently tagged histone modifications on single unfolded chromatin molecules in nanochannels70. c) DNA curtains assay where single DNA molecules are anchored at one end and stretched by fluid flow to image DNA (green) and position quantum dot labeled nucleosomes (magenta)73. (d) Histone oligomers and nucleosomes moving through a solid-state nanopore block different amounts of current (ΔG corresponds to conductance change) and translocate through the nanopore with different times (Δτ)80.
