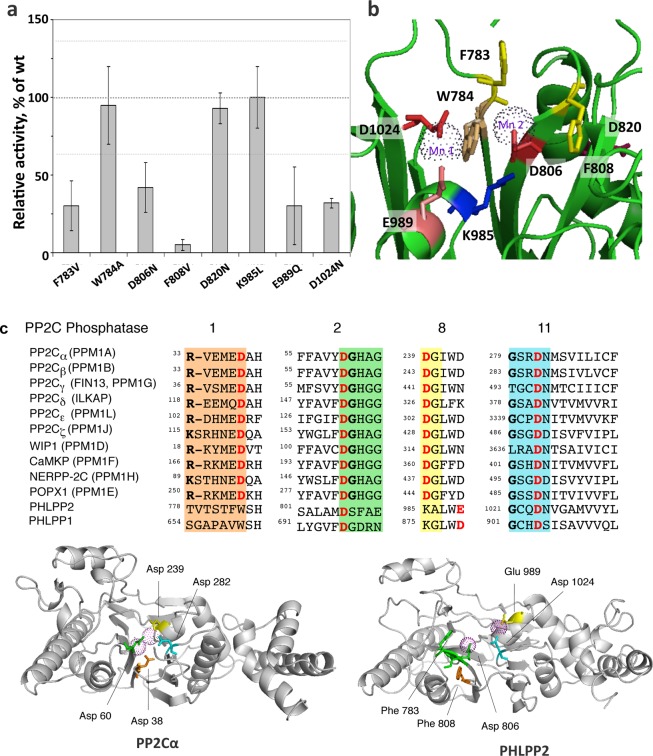
Figure 7.
Mutational analysis of PHLPP2. (a) Relative activity of point mutants of PHLPP2 using a peptide as the substrate. Point mutants of full-length PHLPP2 were overexpressed as HA-tagged fusion proteins in COS7 cells and immunoprecipitated. They were used to dephosphorylate the threonine-phosphorylated peptide RRAPTVA in Tricine (pH 7.5) in the presence of 5 mM MnCl2. Release of inorganic phosphate was monitored by the malachite green assay, and the speed of dephosphorylation was divided by the relative amount of protein, as determined by Western blotting. Activity is given relative to that of wild-type PHLPP2. The graph shows means ± SEM of three separate experiments. (b) Position of the mutated residues in the homology model of PHLPP2.25 (c) Sequence alignment of subdomains 1, 2, 8, and 11 of the PP2C family showing conserved RXXXD, DGXXG, DG, and GXXDN (colored orange, green, yellow, and cyan, respectively). Mn2+-coordinating acidic residues are colored red. Below is a schematic of PP2Cα (left) showing four asparates that coordinate the two bound Mn2+ ions (magenta spheres)34 and a homology model of PHLPP225 showing equivalent acidic residues and structurally important Phe residues.