FIGURE 6:
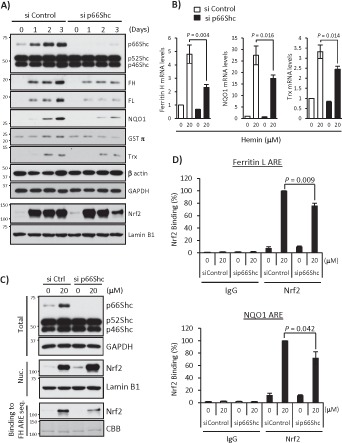
p66Shc deficiency impairs expression of other ARE-dependent antioxidant genes. (A) si Control– or p66Shc siRNA-1–transfected K562 cells were treated with 20 μM hemin for 1, 2, and 3 d. Whole-cell lysates were used in Western blots for Shc, ferritin H (FH), ferritin L (FL), NQO1, GSTπ, thioredoxin-1 (Trx), β-actin, and GAPDH, and nuclear extracts were used for Nrf2 and lamin B1. (B) siRNA-transfected K562 cells were treated with 20 μM hemin for 36 h, and real-time PCR was performed for ferritin H, NQO1, and Trx mRNA. Data are presented as relative mRNA expression (si Control/day 0 is defined as 1.0) after normalization with RPL13A (ribosomal protein L13A). Data are shown as mean ± SD (n = 3). (C) Shc and GAPDH Western blots and Nrf2 and lamin B1 Western blots were performed using whole-cell lysates (Total) and nuclear extracts (Nuc.) of siRNA-transfected K562 cells treated with 20 μM hemin for 36 h, respectively. Nuclear extracts were also used for pull-down assay. Nrf2 binding to biotinylated double-strand ferritin H ARE (−4067 to −4046 base pairs) probe was detected by Western blot using an Nrf2 antibody. Coomassie brilliant blue (CBB) staining is shown for verification of equal loading. (D) siRNA-transfected K562 cells treated with 20 μM hemin for 36 h were subjected to ChIP assays with control IgG or Nrf2 antibody, followed by real-time PCR using ferritin L and NQO1 primers spanning the ARE region. Data were normalized by input DNA and are shown as mean ± SD (n = 3).
