Figure 3.
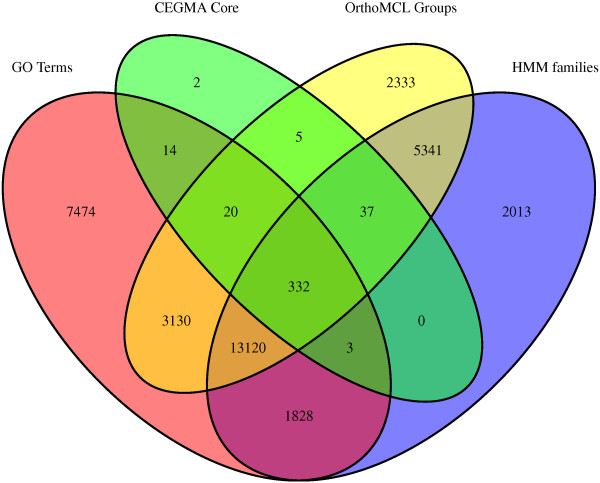
Venn diagram displaying the number of transcripts annotated by each method. Gene Ontology terms were added with Blast2GO using the BLASTX algorithm against NCBI’s nr protein database and a threshold of 1 × 10-03. A set of conserved eukaryotic genes was identified with CEGMA. HMM protein families from the PFAM and TIGR databases were assigned to the amino acid translation of the most likely reading frame for each transcript (identified using an open reading frame prediction tool) using HMMscan under default settings. HMMscan annotations were constrained to a significance threshold of 0.01. Orthogroups were assigned to the same amino acid translations using the orthoMCL web server.
