Figure 4.
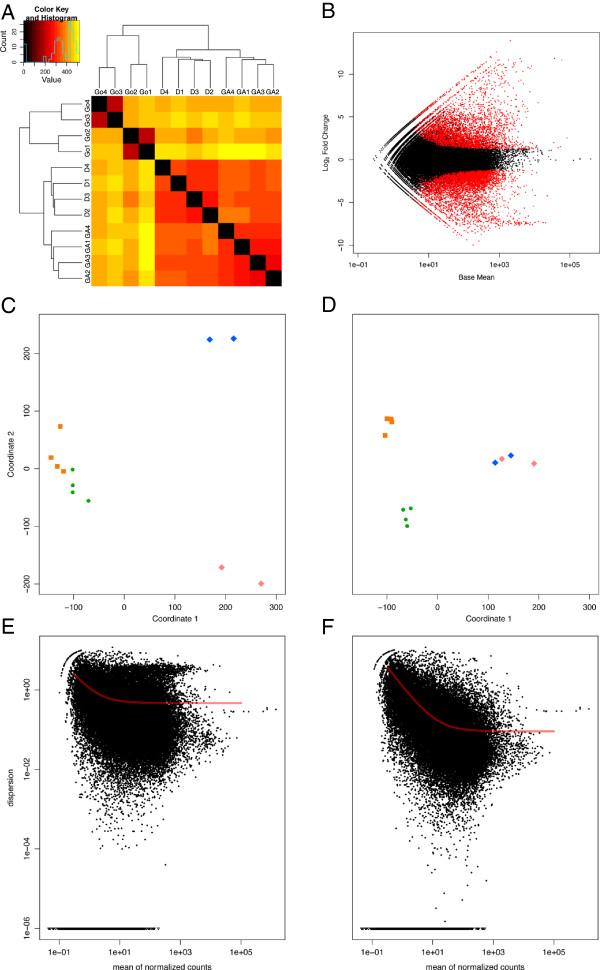
Effects of heterogametic expression on library distances. A. Heatmap of the Euclidean distances between all twelve libraries prior to heterogametic adjustments. Samples Go1 and Go2 correspond to female gonozooid libraries, while Go3 and Go4 correspond to male gonozooid libraries. B. MA plot of the DE analysis between male and female gonozooid libraries in DESeq. Red dots indicate statistically significant DE transcripts. Log2FoldChange > 0 corresponds to expression levels higher in the male gonozooid libraries, and Log2FoldChange < 0 corresponds to expression levels higher in the female gonozooid libraries. C. Euclidean distances plotted in two dimensions prior to heterogametic adjustments. (Legend: orange square-gastrozooids; green circle-dactylozooids; blue diamond-male gonozooids; pink diamond-female gonozooids). D. Euclidean distances plotted in two dimensions after all statistically significant heterogametic transcripts are removed. E. Plot of the estimated dispersion values against the mean of normalized counts of each transcript when binning both male and female gonozooid libraries in a single condition. Fitted dispersion values indicated by the red line. F. Plot of the estimated dispersion values against the mean of normalized counts of each transcript when male and female gonozooid libraries treated as separate conditions.
