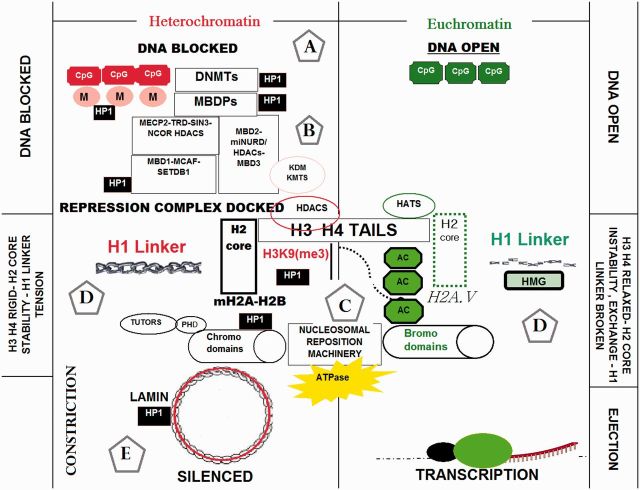
Fig. 1.
Expression of genomic DNA is controlled by heritable epigenetic patterns that result in either transcriptional activated “euchromatin” (right panel) or silenced “heterochromatin” (left panel). [(A) The epigenome diverges from the genome at the level of DNA where CpG sites are methylated by methyltransferases DNMT1, DNMT 3a, and DNMT 3b (which block promoter gene regions to be silenced). (B) Methylated CpGs then attach to methyl-binding proteins (containing methyl-binding domains) “MBDPs” which then recruit diverse transcription repression complexes containing histone modification constriction elements and enzymes such as HDACs and HKMTs. (C) Diverse histone modification enzymes can directly alter histone H3 and H4 tails (tetramers) of the histone unit that corresponds to complementary influences by nucleosomal ATPase reposition machinery containing chromodomains or bromodomains. (D) For gene expression, nucleosomal reposition elements destabilize H2 cores and mobilize nucleosomes through core variant exchange, which initiate nucleosomal ejection, histone H1 linker loss of integrity by several high mobility group proteins (HMG) and a loss of DNA–histone affinity near transcription start sites. In contrast, gene silencing requires H2 core stable variant exchange and recruitment of constrictive nucleosomal remoding complexes, thereby creating mechanical blockage near transcription start sites. (E) Silencing elements are brought together by heterochromatin proteins HP1 α,β which adjoins a number of silencing components in heterochromatin ranging from methylated CpGs, DNMTs, repression complexes such as MBD1-SETDB1-CAF1, silencing marks such as H3K9me3, and nucleosomal reposition machinery. These collective effects reinforce compression of nucleosomes close to the methylated DNA to ensure silencing, when silent heterochromatin is located throughout the lamina circumscribing the nuclear envelope.